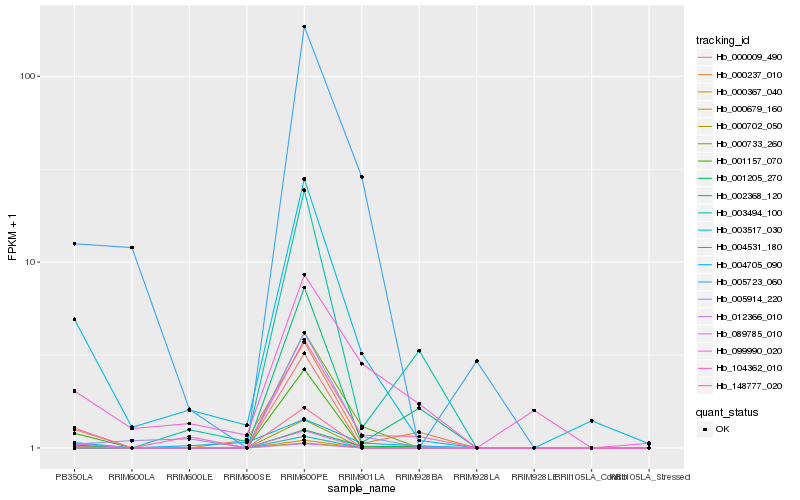
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001205_270 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643859 [Jatropha curcas] |
| 2 |
Hb_000367_040 |
0.1917546339 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 3 |
Hb_004531_180 |
0.1990393796 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 4 |
Hb_001157_070 |
0.2398228346 |
- |
- |
- |
| 5 |
Hb_000009_490 |
0.2404275051 |
- |
- |
hypothetical protein POPTR_0007s12040g [Populus trichocarpa] |
| 6 |
Hb_089785_010 |
0.2451124882 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Fragaria vesca subsp. vesca] |
| 7 |
Hb_012366_010 |
0.2470732132 |
- |
- |
hypothetical protein JCGZ_01894 [Jatropha curcas] |
| 8 |
Hb_005723_060 |
0.2559914865 |
- |
- |
- |
| 9 |
Hb_000702_050 |
0.2728643192 |
transcription factor |
TF Family: ERF |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_099990_020 |
0.2823964463 |
- |
- |
hypothetical protein POPTR_0006s28940g [Populus trichocarpa] |
| 11 |
Hb_003517_030 |
0.2830942346 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 5 isoform X2 [Eucalyptus grandis] |
| 12 |
Hb_005914_220 |
0.2846264327 |
transcription factor |
TF Family: Orphans |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004705_090 |
0.2848256553 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_104362_010 |
0.2857670848 |
- |
- |
hypothetical protein POPTR_0247s002302g, partial [Populus trichocarpa] |
| 15 |
Hb_000679_160 |
0.286466494 |
- |
- |
PREDICTED: probable carboxylesterase 17 [Jatropha curcas] |
| 16 |
Hb_002368_120 |
0.2879493022 |
- |
- |
hypothetical protein VITISV_033181 [Vitis vinifera] |
| 17 |
Hb_000733_260 |
0.2898335004 |
- |
- |
hypothetical protein JCGZ_26934 [Jatropha curcas] |
| 18 |
Hb_148777_020 |
0.2957375822 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11410 isoform X1 [Populus euphratica] |
| 19 |
Hb_003494_100 |
0.2958118037 |
transcription factor |
TF Family: zf-HD |
Mini zinc finger 2 isoform 1 [Theobroma cacao] |
| 20 |
Hb_000237_010 |
0.2963117088 |
transcription factor |
TF Family: SRS |
conserved hypothetical protein [Ricinus communis] |