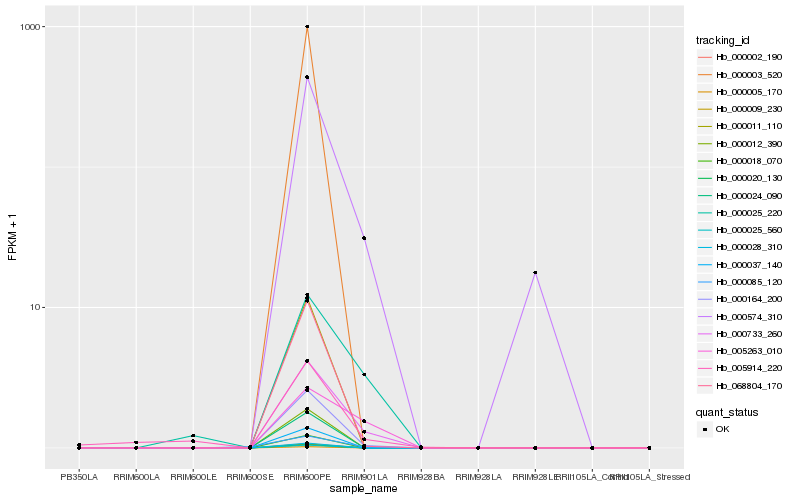
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000733_260 |
0.0 |
- |
- |
hypothetical protein JCGZ_26934 [Jatropha curcas] |
| 2 |
Hb_000164_200 |
0.1185766457 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 3 |
Hb_000574_310 |
0.1375820937 |
- |
- |
hypothetical protein CISIN_1g027539mg [Citrus sinensis] |
| 4 |
Hb_000025_220 |
0.1380987131 |
- |
- |
hypothetical protein POPTR_0001s28080g [Populus trichocarpa] |
| 5 |
Hb_005263_010 |
0.1825809238 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At2g33170 [Nicotiana sylvestris] |
| 6 |
Hb_068804_170 |
0.1941562436 |
- |
- |
PREDICTED: probable rhamnogalacturonate lyase B [Jatropha curcas] |
| 7 |
Hb_000085_120 |
0.1954236811 |
- |
- |
Benzoate carboxyl methyltransferase, putative [Ricinus communis] |
| 8 |
Hb_005914_220 |
0.2109836418 |
transcription factor |
TF Family: Orphans |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000002_190 |
0.2128792366 |
transcription factor |
TF Family: ERF |
Ethylene-responsive transcription factor [Morus notabilis] |
| 10 |
Hb_000003_520 |
0.2128792366 |
- |
- |
PREDICTED: cell division cycle protein 27 homolog B [Jatropha curcas] |
| 11 |
Hb_000005_170 |
0.2128792366 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR [Jatropha curcas] |
| 12 |
Hb_000009_230 |
0.2128792366 |
- |
- |
hypothetical protein POPTR_0007s12290g [Populus trichocarpa] |
| 13 |
Hb_000011_110 |
0.2128792366 |
- |
- |
PREDICTED: uncharacterized protein LOC105631299 [Jatropha curcas] |
| 14 |
Hb_000012_390 |
0.2128792366 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000018_070 |
0.2128792366 |
- |
- |
PREDICTED: amino acid permease 8-like [Jatropha curcas] |
| 16 |
Hb_000020_130 |
0.2128792366 |
- |
- |
conidial pigment biosynthesis oxidase Arb2/brown2 [Aspergillus clavatus NRRL 1] |
| 17 |
Hb_000024_090 |
0.2128792366 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000025_560 |
0.2128792366 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 19 |
Hb_000028_310 |
0.2128792366 |
- |
- |
PREDICTED: homeobox protein knotted-1-like 1 [Jatropha curcas] |
| 20 |
Hb_000037_140 |
0.2128792366 |
- |
- |
PREDICTED: uncharacterized protein LOC104586896 [Nelumbo nucifera] |