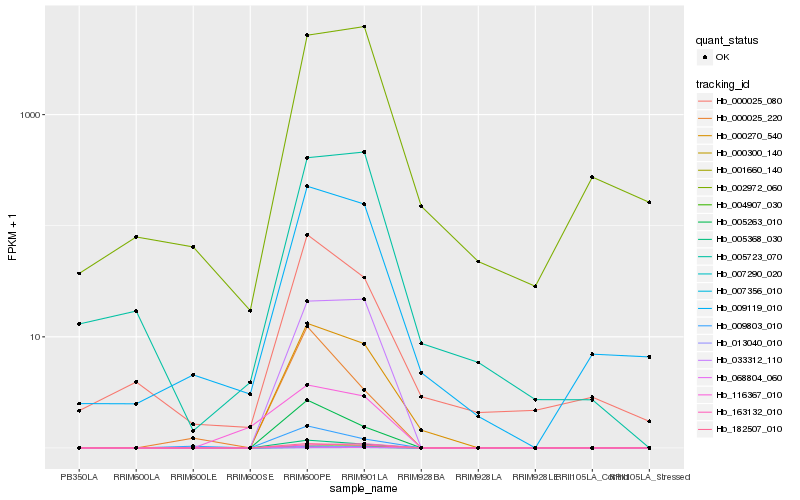
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001660_140 |
0.0 |
- |
- |
- |
| 2 |
Hb_182507_010 |
0.0033637657 |
- |
- |
- |
| 3 |
Hb_013040_010 |
0.0219288904 |
- |
- |
hypothetical protein POPTR_0009s09690g [Populus trichocarpa] |
| 4 |
Hb_005368_030 |
0.0622437936 |
- |
- |
- |
| 5 |
Hb_004907_030 |
0.0814941706 |
- |
- |
hypothetical protein POPTR_0012s032802g, partial [Populus trichocarpa] |
| 6 |
Hb_000270_540 |
0.1049768514 |
- |
- |
hypothetical protein JCGZ_11527 [Jatropha curcas] |
| 7 |
Hb_033312_110 |
0.1057806412 |
- |
- |
- |
| 8 |
Hb_005263_010 |
0.129776734 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At2g33170 [Nicotiana sylvestris] |
| 9 |
Hb_000300_140 |
0.1537035449 |
transcription factor |
TF Family: HB |
Protein WUSCHEL, putative [Ricinus communis] |
| 10 |
Hb_068804_060 |
0.1594361914 |
- |
- |
PREDICTED: ABC transporter G family member 17-like [Jatropha curcas] |
| 11 |
Hb_007290_020 |
0.159582511 |
- |
- |
PREDICTED: subtilisin-like protease SBT4.14 [Jatropha curcas] |
| 12 |
Hb_007356_010 |
0.164845394 |
- |
- |
CTV.19 [Citrus trifoliata] |
| 13 |
Hb_163132_010 |
0.1690148375 |
- |
- |
- |
| 14 |
Hb_009119_010 |
0.1778626444 |
- |
- |
- |
| 15 |
Hb_009803_010 |
0.188312963 |
- |
- |
hypothetical protein JCGZ_08257 [Jatropha curcas] |
| 16 |
Hb_005723_070 |
0.2022720763 |
- |
- |
hypothetical protein CISIN_1g011514mg [Citrus sinensis] |
| 17 |
Hb_000025_220 |
0.2254363697 |
- |
- |
hypothetical protein POPTR_0001s28080g [Populus trichocarpa] |
| 18 |
Hb_002972_060 |
0.2306351071 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 19 |
Hb_116367_010 |
0.2354489532 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Pyrus x bretschneideri] |
| 20 |
Hb_000025_080 |
0.2375190162 |
- |
- |
- |