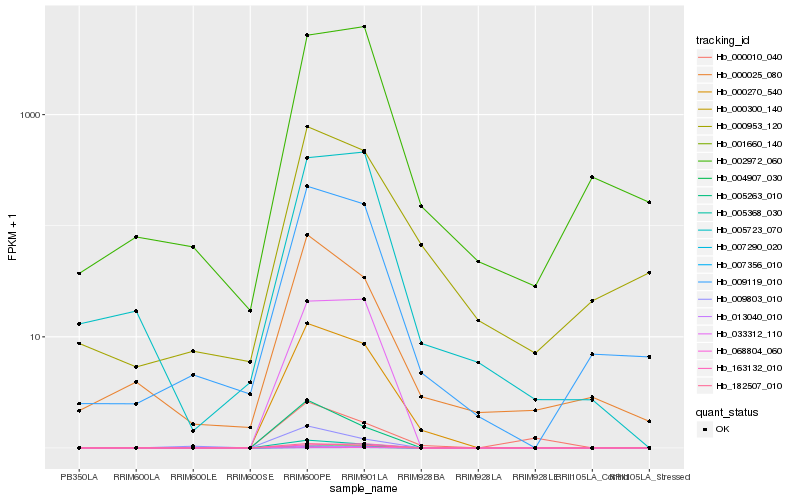
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000270_540 |
0.0 |
- |
- |
hypothetical protein JCGZ_11527 [Jatropha curcas] |
| 2 |
Hb_001660_140 |
0.1049768514 |
- |
- |
- |
| 3 |
Hb_182507_010 |
0.1049978459 |
- |
- |
- |
| 4 |
Hb_013040_010 |
0.107423836 |
- |
- |
hypothetical protein POPTR_0009s09690g [Populus trichocarpa] |
| 5 |
Hb_005368_030 |
0.1213545405 |
- |
- |
- |
| 6 |
Hb_004907_030 |
0.1332339226 |
- |
- |
hypothetical protein POPTR_0012s032802g, partial [Populus trichocarpa] |
| 7 |
Hb_033312_110 |
0.1493227924 |
- |
- |
- |
| 8 |
Hb_005263_010 |
0.1656006094 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At2g33170 [Nicotiana sylvestris] |
| 9 |
Hb_009119_010 |
0.16893657 |
- |
- |
- |
| 10 |
Hb_000300_140 |
0.1862512801 |
transcription factor |
TF Family: HB |
Protein WUSCHEL, putative [Ricinus communis] |
| 11 |
Hb_068804_060 |
0.1909872887 |
- |
- |
PREDICTED: ABC transporter G family member 17-like [Jatropha curcas] |
| 12 |
Hb_007290_020 |
0.1911088604 |
- |
- |
PREDICTED: subtilisin-like protease SBT4.14 [Jatropha curcas] |
| 13 |
Hb_007356_010 |
0.1955033671 |
- |
- |
CTV.19 [Citrus trifoliata] |
| 14 |
Hb_005723_070 |
0.1966487555 |
- |
- |
hypothetical protein CISIN_1g011514mg [Citrus sinensis] |
| 15 |
Hb_163132_010 |
0.1990137125 |
- |
- |
- |
| 16 |
Hb_000953_120 |
0.2006281204 |
- |
- |
- |
| 17 |
Hb_009803_010 |
0.2139299092 |
- |
- |
hypothetical protein JCGZ_08257 [Jatropha curcas] |
| 18 |
Hb_002972_060 |
0.2184982902 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 19 |
Hb_000025_080 |
0.2202834766 |
- |
- |
- |
| 20 |
Hb_000010_040 |
0.2259517478 |
- |
- |
PREDICTED: probable 1-aminocyclopropane-1-carboxylate deaminase-like isoform X2 [Citrus sinensis] |