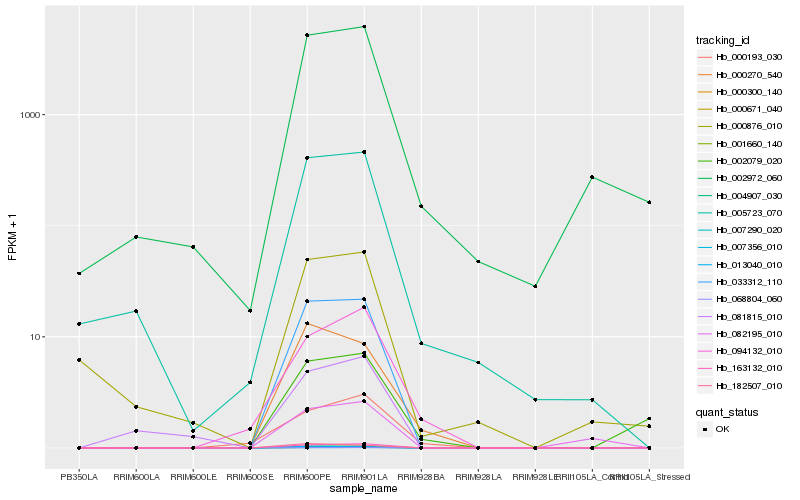
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007290_020 |
0.0 |
- |
- |
PREDICTED: subtilisin-like protease SBT4.14 [Jatropha curcas] |
| 2 |
Hb_068804_060 |
0.0001476233 |
- |
- |
PREDICTED: ABC transporter G family member 17-like [Jatropha curcas] |
| 3 |
Hb_007356_010 |
0.0053114054 |
- |
- |
CTV.19 [Citrus trifoliata] |
| 4 |
Hb_000300_140 |
0.0059294305 |
transcription factor |
TF Family: HB |
Protein WUSCHEL, putative [Ricinus communis] |
| 5 |
Hb_163132_010 |
0.0095215461 |
- |
- |
- |
| 6 |
Hb_033312_110 |
0.0541188506 |
- |
- |
- |
| 7 |
Hb_004907_030 |
0.0784430894 |
- |
- |
hypothetical protein POPTR_0012s032802g, partial [Populus trichocarpa] |
| 8 |
Hb_013040_010 |
0.137823311 |
- |
- |
hypothetical protein POPTR_0009s09690g [Populus trichocarpa] |
| 9 |
Hb_000671_040 |
0.1406808625 |
- |
- |
- |
| 10 |
Hb_001660_140 |
0.159582511 |
- |
- |
- |
| 11 |
Hb_182507_010 |
0.162915273 |
- |
- |
- |
| 12 |
Hb_005723_070 |
0.1668159381 |
- |
- |
hypothetical protein CISIN_1g011514mg [Citrus sinensis] |
| 13 |
Hb_094132_010 |
0.1699895092 |
- |
- |
hypothetical protein POPTR_0018s03360g [Populus trichocarpa] |
| 14 |
Hb_000193_030 |
0.1820732093 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Jatropha curcas] |
| 15 |
Hb_081815_010 |
0.1868242467 |
- |
- |
PREDICTED: uncharacterized protein LOC100266863 [Vitis vinifera] |
| 16 |
Hb_082195_010 |
0.1890864629 |
- |
- |
Uncharacterized protein TCM_043808 [Theobroma cacao] |
| 17 |
Hb_002972_060 |
0.1904751723 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 18 |
Hb_000270_540 |
0.1911088604 |
- |
- |
hypothetical protein JCGZ_11527 [Jatropha curcas] |
| 19 |
Hb_000876_010 |
0.2080680203 |
- |
- |
Uncharacterized protein TCM_022272 [Theobroma cacao] |
| 20 |
Hb_002079_020 |
0.2099829166 |
- |
- |
- |