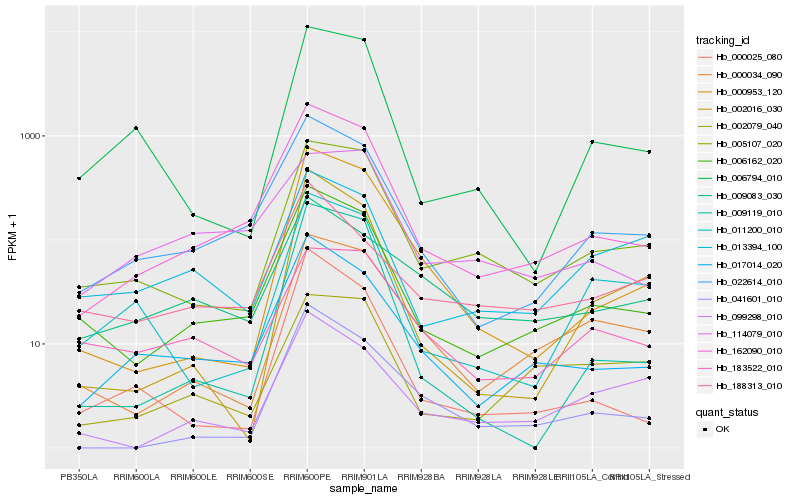
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_162090_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642032 [Jatropha curcas] |
| 2 |
Hb_022614_010 |
0.0841465473 |
- |
- |
- |
| 3 |
Hb_006162_020 |
0.0912595885 |
- |
- |
- |
| 4 |
Hb_017014_020 |
0.1148914198 |
- |
- |
- |
| 5 |
Hb_005107_020 |
0.1539241311 |
- |
- |
hypothetical protein JCGZ_25266 [Jatropha curcas] |
| 6 |
Hb_000953_120 |
0.1649059722 |
- |
- |
- |
| 7 |
Hb_041601_010 |
0.1712195861 |
- |
- |
- |
| 8 |
Hb_000025_080 |
0.1720519176 |
- |
- |
- |
| 9 |
Hb_000034_090 |
0.1759725961 |
transcription factor |
TF Family: M-type |
MPF2-like, partial [Dunalia fasciculata] |
| 10 |
Hb_009119_010 |
0.1771591316 |
- |
- |
- |
| 11 |
Hb_006794_010 |
0.1873534005 |
- |
- |
elicitor-responsive protein [Hevea brasiliensis] |
| 12 |
Hb_114079_010 |
0.1881121078 |
- |
- |
hypothetical protein CICLE_v10008147mg [Citrus clementina] |
| 13 |
Hb_011200_010 |
0.1942878879 |
- |
- |
- |
| 14 |
Hb_002016_030 |
0.1979693063 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_009083_030 |
0.2021957386 |
- |
- |
hypothetical protein POPTR_0007s14760g [Populus trichocarpa] |
| 16 |
Hb_183522_010 |
0.2063145712 |
- |
- |
Far upstream element-binding protein, putative [Ricinus communis] |
| 17 |
Hb_099298_010 |
0.2104808391 |
- |
- |
- |
| 18 |
Hb_002079_040 |
0.2135454176 |
- |
- |
hypothetical protein VITISV_018542 [Vitis vinifera] |
| 19 |
Hb_188313_010 |
0.2156769965 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 20 |
Hb_013394_100 |
0.2160122493 |
- |
- |
PREDICTED: uncharacterized protein LOC105135144 [Populus euphratica] |