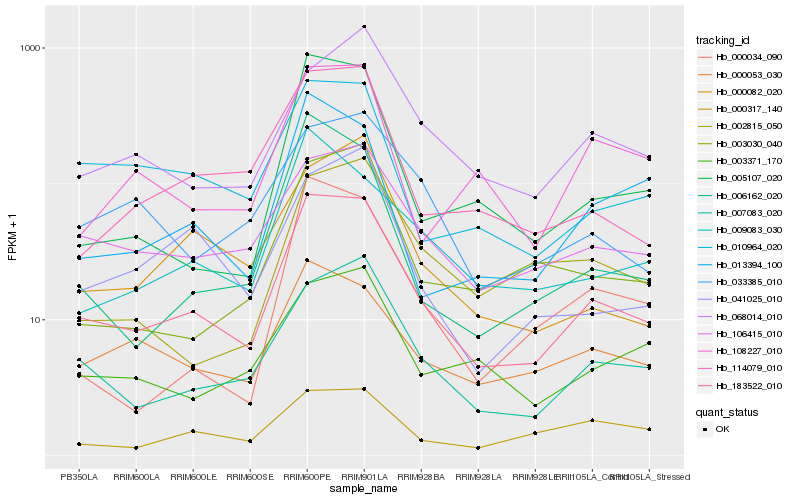
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_183522_010 |
0.0 |
- |
- |
Far upstream element-binding protein, putative [Ricinus communis] |
| 2 |
Hb_007083_020 |
0.1249472777 |
- |
- |
hypothetical protein F775_15403 [Aegilops tauschii] |
| 3 |
Hb_106415_010 |
0.1440654648 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X1 [Jatropha curcas] |
| 4 |
Hb_114079_010 |
0.152723439 |
- |
- |
hypothetical protein CICLE_v10008147mg [Citrus clementina] |
| 5 |
Hb_010964_020 |
0.1568856685 |
- |
- |
RNA helicase [Arabidopsis thaliana] |
| 6 |
Hb_000053_030 |
0.1585600851 |
- |
- |
hypothetical protein CISIN_1g0010393mg, partial [Citrus sinensis] |
| 7 |
Hb_005107_020 |
0.1615964908 |
- |
- |
hypothetical protein JCGZ_25266 [Jatropha curcas] |
| 8 |
Hb_009083_030 |
0.1665294926 |
- |
- |
hypothetical protein POPTR_0007s14760g [Populus trichocarpa] |
| 9 |
Hb_033385_010 |
0.1675831213 |
- |
- |
hypothetical protein JCGZ_06094 [Jatropha curcas] |
| 10 |
Hb_000082_020 |
0.1705124102 |
- |
- |
- |
| 11 |
Hb_003030_040 |
0.1705259734 |
- |
- |
hypothetical protein CISIN_1g0466212mg, partial [Citrus sinensis] |
| 12 |
Hb_006162_020 |
0.1708976555 |
- |
- |
- |
| 13 |
Hb_041025_010 |
0.1713311376 |
- |
- |
PREDICTED: uncharacterized protein LOC105635593 [Jatropha curcas] |
| 14 |
Hb_068014_010 |
0.1722111475 |
- |
- |
hypothetical protein POPTR_0312s00200g, partial [Populus trichocarpa] |
| 15 |
Hb_003371_170 |
0.1736692949 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_013394_100 |
0.1737783329 |
- |
- |
PREDICTED: uncharacterized protein LOC105135144 [Populus euphratica] |
| 17 |
Hb_108227_010 |
0.1759624049 |
- |
- |
RecName: Full=Profilin-2; AltName: Full=Pollen allergen Hev b 8.0102; AltName: Allergen=Hev b 8.0102 [Hevea brasiliensis] |
| 18 |
Hb_000034_090 |
0.1762443684 |
transcription factor |
TF Family: M-type |
MPF2-like, partial [Dunalia fasciculata] |
| 19 |
Hb_002815_050 |
0.1804960229 |
- |
- |
PREDICTED: succinate-semialdehyde dehydrogenase, mitochondrial [Populus euphratica] |
| 20 |
Hb_000317_140 |
0.1819428463 |
- |
- |
hypothetical protein CISIN_1g0019781mg, partial [Citrus sinensis] |