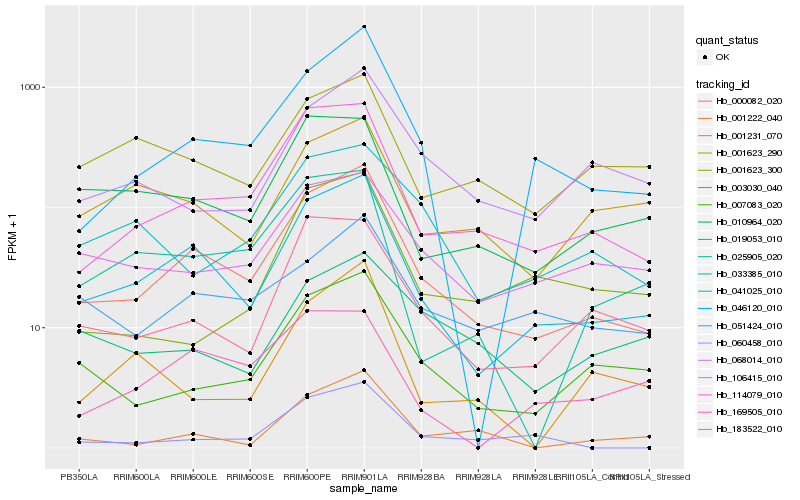
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000082_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_041025_010 |
0.0892649558 |
- |
- |
PREDICTED: uncharacterized protein LOC105635593 [Jatropha curcas] |
| 3 |
Hb_114079_010 |
0.1257546855 |
- |
- |
hypothetical protein CICLE_v10008147mg [Citrus clementina] |
| 4 |
Hb_046120_010 |
0.1515699447 |
- |
- |
- |
| 5 |
Hb_007083_020 |
0.1696047053 |
- |
- |
hypothetical protein F775_15403 [Aegilops tauschii] |
| 6 |
Hb_183522_010 |
0.1705124102 |
- |
- |
Far upstream element-binding protein, putative [Ricinus communis] |
| 7 |
Hb_106415_010 |
0.1883702965 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X1 [Jatropha curcas] |
| 8 |
Hb_051424_010 |
0.1922950921 |
- |
- |
PREDICTED: putative methyltransferase At1g22800 [Jatropha curcas] |
| 9 |
Hb_033385_010 |
0.1940226139 |
- |
- |
hypothetical protein JCGZ_06094 [Jatropha curcas] |
| 10 |
Hb_068014_010 |
0.194069854 |
- |
- |
hypothetical protein POPTR_0312s00200g, partial [Populus trichocarpa] |
| 11 |
Hb_025905_020 |
0.1961672997 |
- |
- |
major allergen Pru ar 1-like [Jatropha curcas] |
| 12 |
Hb_010964_020 |
0.1963140052 |
- |
- |
RNA helicase [Arabidopsis thaliana] |
| 13 |
Hb_001222_040 |
0.1963853454 |
- |
- |
- |
| 14 |
Hb_060458_010 |
0.202290442 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001623_290 |
0.2083423595 |
- |
- |
unknown [Populus trichocarpa] |
| 16 |
Hb_001231_070 |
0.2108009698 |
- |
- |
- |
| 17 |
Hb_003030_040 |
0.211136797 |
- |
- |
hypothetical protein CISIN_1g0466212mg, partial [Citrus sinensis] |
| 18 |
Hb_001623_300 |
0.2120185911 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13-B [Jatropha curcas] |
| 19 |
Hb_019053_010 |
0.2172244211 |
- |
- |
- |
| 20 |
Hb_169505_010 |
0.2177223144 |
- |
- |
hypothetical protein JCGZ_04480 [Jatropha curcas] |