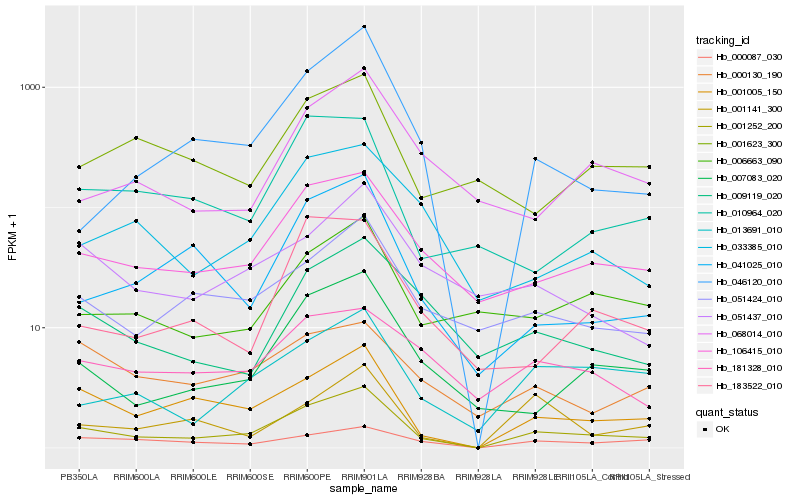
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001252_200 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640649 [Jatropha curcas] |
| 2 |
Hb_001005_150 |
0.1418362917 |
- |
- |
- |
| 3 |
Hb_106415_010 |
0.1560815045 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X1 [Jatropha curcas] |
| 4 |
Hb_007083_020 |
0.1759907193 |
- |
- |
hypothetical protein F775_15403 [Aegilops tauschii] |
| 5 |
Hb_051424_010 |
0.1821358433 |
- |
- |
PREDICTED: putative methyltransferase At1g22800 [Jatropha curcas] |
| 6 |
Hb_013691_010 |
0.1838604158 |
- |
- |
PREDICTED: uncharacterized protein LOC103724099, partial [Phoenix dactylifera] |
| 7 |
Hb_000087_030 |
0.1946221787 |
- |
- |
hypothetical protein JCGZ_02243 [Jatropha curcas] |
| 8 |
Hb_006663_090 |
0.1979068041 |
- |
- |
hypothetical protein B456_010G1255001, partial [Gossypium raimondii] |
| 9 |
Hb_041025_010 |
0.2009048781 |
- |
- |
PREDICTED: uncharacterized protein LOC105635593 [Jatropha curcas] |
| 10 |
Hb_033385_010 |
0.2013485615 |
- |
- |
hypothetical protein JCGZ_06094 [Jatropha curcas] |
| 11 |
Hb_046120_010 |
0.2021227315 |
- |
- |
- |
| 12 |
Hb_009119_020 |
0.2039926036 |
- |
- |
hypothetical protein CISIN_1g033708mg [Citrus sinensis] |
| 13 |
Hb_001141_300 |
0.2066556934 |
- |
- |
- |
| 14 |
Hb_051437_010 |
0.2087258893 |
- |
- |
PREDICTED: calmodulin-interacting protein 111 isoform X2 [Jatropha curcas] |
| 15 |
Hb_010964_020 |
0.2098890972 |
- |
- |
RNA helicase [Arabidopsis thaliana] |
| 16 |
Hb_181328_010 |
0.2106792947 |
- |
- |
PREDICTED: ferredoxin-dependent glutamate synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_183522_010 |
0.2118052236 |
- |
- |
Far upstream element-binding protein, putative [Ricinus communis] |
| 18 |
Hb_068014_010 |
0.211952724 |
- |
- |
hypothetical protein POPTR_0312s00200g, partial [Populus trichocarpa] |
| 19 |
Hb_000130_190 |
0.213337593 |
- |
- |
PREDICTED: uncharacterized protein LOC105638437 [Jatropha curcas] |
| 20 |
Hb_001623_300 |
0.2153979469 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13-B [Jatropha curcas] |