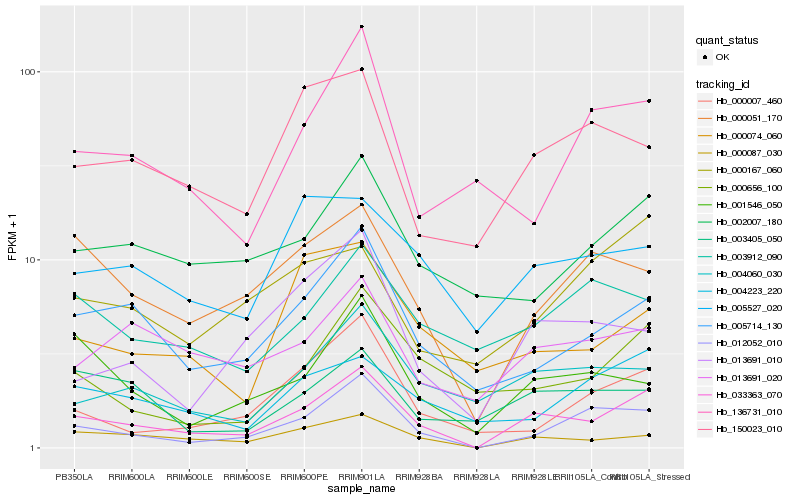
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033363_070 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: methionine aminopeptidase 1B, chloroplastic, partial [Populus euphratica] |
| 2 |
Hb_000087_030 |
0.1566993737 |
- |
- |
hypothetical protein JCGZ_02243 [Jatropha curcas] |
| 3 |
Hb_012052_010 |
0.1674430758 |
- |
- |
PREDICTED: uncharacterized protein LOC103323653 [Prunus mume] |
| 4 |
Hb_005714_130 |
0.1818479309 |
transcription factor |
TF Family: C2H2 |
unknown [Populus trichocarpa x Populus deltoides] |
| 5 |
Hb_000051_170 |
0.2017757555 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 6 |
Hb_000656_100 |
0.2027198978 |
- |
- |
PREDICTED: uncharacterized protein LOC105633651 [Jatropha curcas] |
| 7 |
Hb_004060_030 |
0.2028458216 |
- |
- |
PREDICTED: primase homolog protein [Jatropha curcas] |
| 8 |
Hb_002007_180 |
0.2041726546 |
- |
- |
PREDICTED: uncharacterized protein LOC105647525 [Jatropha curcas] |
| 9 |
Hb_013691_020 |
0.2092698405 |
- |
- |
transcription initiation factor iie, alpha subunit, putative [Ricinus communis] |
| 10 |
Hb_001546_050 |
0.2105405624 |
- |
- |
- |
| 11 |
Hb_004223_220 |
0.2113579705 |
- |
- |
PREDICTED: uncharacterized protein LOC105635366 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000007_460 |
0.2152518435 |
- |
- |
PREDICTED: uncharacterized protein LOC105641948 [Jatropha curcas] |
| 13 |
Hb_013691_010 |
0.2154998866 |
- |
- |
PREDICTED: uncharacterized protein LOC103724099, partial [Phoenix dactylifera] |
| 14 |
Hb_150023_010 |
0.2163271337 |
- |
- |
50S ribosomal protein L2, putative [Ricinus communis] |
| 15 |
Hb_005527_020 |
0.2182241185 |
- |
- |
PREDICTED: uncharacterized protein LOC105643027 [Jatropha curcas] |
| 16 |
Hb_000074_060 |
0.2234805642 |
- |
- |
PREDICTED: membrane-associated progesterone-binding protein 4-like [Populus euphratica] |
| 17 |
Hb_136731_010 |
0.2254146778 |
- |
- |
PREDICTED: uncharacterized protein LOC105628378 isoform X4 [Jatropha curcas] |
| 18 |
Hb_003912_090 |
0.2254467868 |
- |
- |
peptidyl-tRNA hydrolase, putative [Ricinus communis] |
| 19 |
Hb_000167_060 |
0.2270312979 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 31 kDa protein [Jatropha curcas] |
| 20 |
Hb_003405_050 |
0.227181909 |
- |
- |
- |