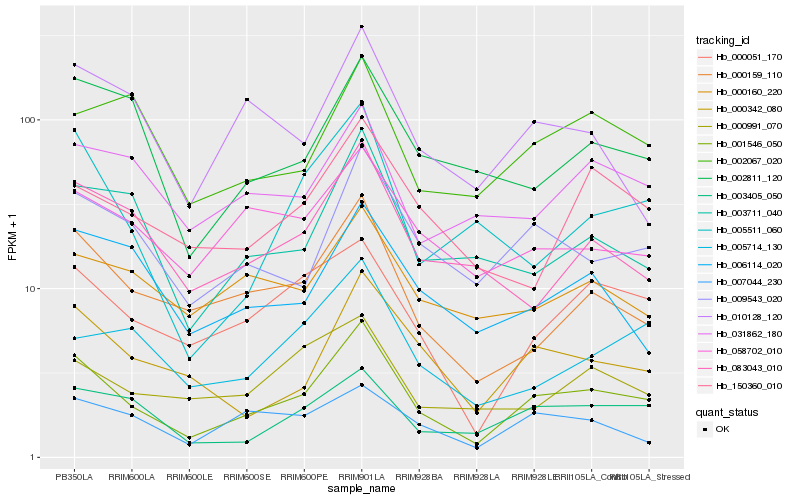
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001546_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_000159_110 |
0.1489847057 |
- |
- |
PREDICTED: thioredoxin-like protein YLS8 isoform X2 [Malus domestica] |
| 3 |
Hb_009543_020 |
0.1494972761 |
- |
- |
PREDICTED: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 4 |
Hb_000051_170 |
0.1520681791 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 5 |
Hb_000342_080 |
0.1542686765 |
- |
- |
hypothetical protein L484_019524 [Morus notabilis] |
| 6 |
Hb_003405_050 |
0.158494358 |
- |
- |
- |
| 7 |
Hb_003711_040 |
0.172402205 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Jatropha curcas] |
| 8 |
Hb_083043_010 |
0.175442464 |
- |
- |
PREDICTED: ribonuclease P protein subunit p25-like protein [Jatropha curcas] |
| 9 |
Hb_058702_010 |
0.1754545914 |
- |
- |
PREDICTED: importin beta-like SAD2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002811_120 |
0.1792965708 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT102 [Jatropha curcas] |
| 11 |
Hb_031862_180 |
0.1800354874 |
- |
- |
PREDICTED: uncharacterized protein LOC105644669 [Jatropha curcas] |
| 12 |
Hb_006114_020 |
0.1807463406 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005511_060 |
0.1809345258 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_007044_230 |
0.1814877592 |
- |
- |
PREDICTED: uncharacterized protein LOC105638702 [Jatropha curcas] |
| 15 |
Hb_010128_120 |
0.1817962782 |
- |
- |
hypothetical protein JCGZ_23167 [Jatropha curcas] |
| 16 |
Hb_000160_220 |
0.1820356212 |
- |
- |
PREDICTED: protein AAR2 homolog isoform X1 [Jatropha curcas] |
| 17 |
Hb_005714_130 |
0.182146675 |
transcription factor |
TF Family: C2H2 |
unknown [Populus trichocarpa x Populus deltoides] |
| 18 |
Hb_150360_010 |
0.1821724885 |
- |
- |
hypothetical protein JCGZ_16890 [Jatropha curcas] |
| 19 |
Hb_000991_070 |
0.1826888936 |
- |
- |
phytochrome A specific signal transduction component family protein [Populus trichocarpa] |
| 20 |
Hb_002067_020 |
0.182857262 |
- |
- |
conserved hypothetical protein [Ricinus communis] |