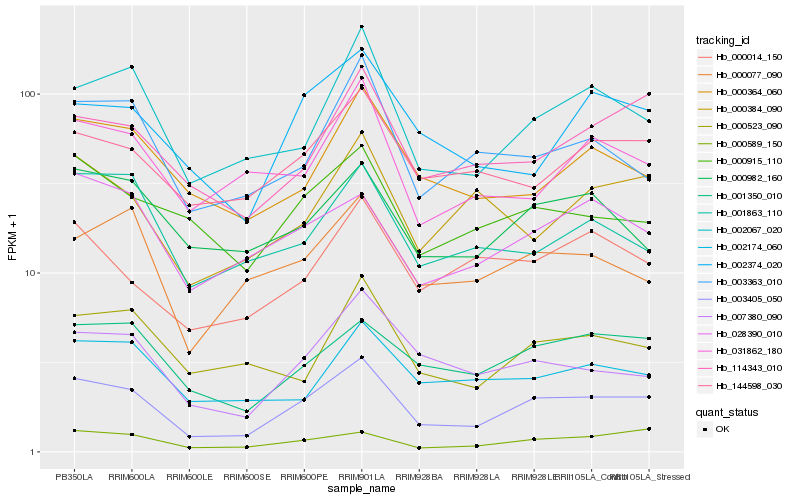
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003405_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_002067_020 |
0.1204124098 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001350_010 |
0.1254026892 |
- |
- |
PREDICTED: uncharacterized protein LOC105629383 [Jatropha curcas] |
| 4 |
Hb_000589_150 |
0.1293906082 |
- |
- |
PREDICTED: factor of DNA methylation 1-like isoform X1 [Beta vulgaris subsp. vulgaris] |
| 5 |
Hb_007380_090 |
0.131914735 |
- |
- |
- |
| 6 |
Hb_114343_010 |
0.1360840359 |
- |
- |
PREDICTED: uncharacterized protein LOC105647831 [Jatropha curcas] |
| 7 |
Hb_003363_010 |
0.1375398692 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_144598_030 |
0.1386096681 |
- |
- |
PREDICTED: mitotic apparatus protein p62 [Jatropha curcas] |
| 9 |
Hb_028390_010 |
0.1407908922 |
- |
- |
PREDICTED: filament-like plant protein 3 [Populus euphratica] |
| 10 |
Hb_000077_090 |
0.1410592046 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000915_110 |
0.1417675786 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase A5 [Jatropha curcas] |
| 12 |
Hb_000364_060 |
0.1419002597 |
- |
- |
JHL03K20.4 [Jatropha curcas] |
| 13 |
Hb_000523_090 |
0.1428723448 |
- |
- |
PREDICTED: protein DJ-1 homolog D-like [Populus euphratica] |
| 14 |
Hb_000982_160 |
0.1434883088 |
- |
- |
PREDICTED: CLIP-associated protein [Jatropha curcas] |
| 15 |
Hb_002374_020 |
0.1443289787 |
- |
- |
PREDICTED: protein KRTCAP2 homolog [Jatropha curcas] |
| 16 |
Hb_002174_060 |
0.1450371063 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g09900-like [Jatropha curcas] |
| 17 |
Hb_001863_110 |
0.1462137314 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 18 |
Hb_031862_180 |
0.1469166424 |
- |
- |
PREDICTED: uncharacterized protein LOC105644669 [Jatropha curcas] |
| 19 |
Hb_000014_150 |
0.1471634397 |
- |
- |
PREDICTED: cysteine--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 20 |
Hb_000384_090 |
0.1474082385 |
- |
- |
PREDICTED: probable VAMP-like protein At1g33475 [Jatropha curcas] |