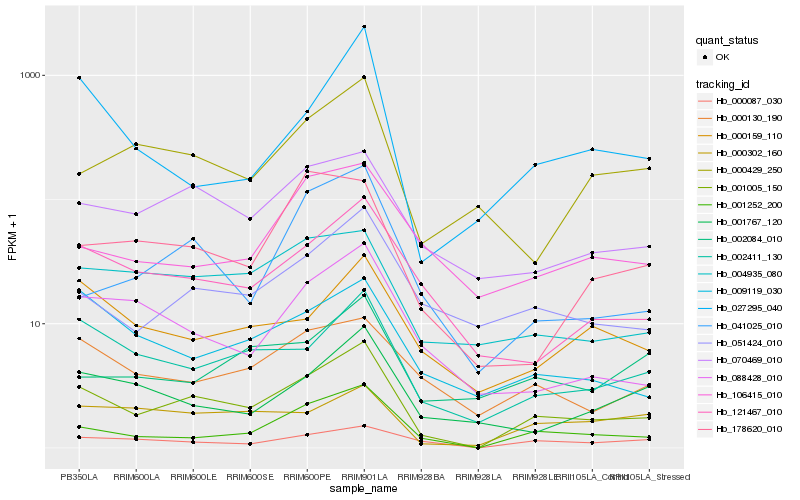
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001005_150 |
0.0 |
- |
- |
- |
| 2 |
Hb_001252_200 |
0.1418362917 |
- |
- |
PREDICTED: uncharacterized protein LOC105640649 [Jatropha curcas] |
| 3 |
Hb_002411_130 |
0.1673036822 |
- |
- |
PREDICTED: cytosolic sulfotransferase 15-like [Populus euphratica] |
| 4 |
Hb_070469_010 |
0.1801766361 |
- |
- |
hypothetical protein POPTR_0002s21850g [Populus trichocarpa] |
| 5 |
Hb_121467_010 |
0.182845349 |
- |
- |
tubulin-specific chaperone E, putative [Ricinus communis] |
| 6 |
Hb_051424_010 |
0.1867084337 |
- |
- |
PREDICTED: putative methyltransferase At1g22800 [Jatropha curcas] |
| 7 |
Hb_000087_030 |
0.1924648156 |
- |
- |
hypothetical protein JCGZ_02243 [Jatropha curcas] |
| 8 |
Hb_004935_080 |
0.1961222304 |
- |
- |
AT3g13060/MGH6_17 [Arabidopsis thaliana] |
| 9 |
Hb_000130_190 |
0.1985022263 |
- |
- |
PREDICTED: uncharacterized protein LOC105638437 [Jatropha curcas] |
| 10 |
Hb_000159_110 |
0.1997283963 |
- |
- |
PREDICTED: thioredoxin-like protein YLS8 isoform X2 [Malus domestica] |
| 11 |
Hb_088428_010 |
0.2020188801 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181 homolog [Jatropha curcas] |
| 12 |
Hb_009119_030 |
0.2032466116 |
- |
- |
GMFP4 [Medicago truncatula] |
| 13 |
Hb_000302_160 |
0.2045509284 |
- |
- |
PREDICTED: uncharacterized protein LOC105648403 [Jatropha curcas] |
| 14 |
Hb_000429_250 |
0.2078477778 |
- |
- |
hypothetical protein L484_000744 [Morus notabilis] |
| 15 |
Hb_001767_120 |
0.2083011361 |
- |
- |
- |
| 16 |
Hb_041025_010 |
0.2141556502 |
- |
- |
PREDICTED: uncharacterized protein LOC105635593 [Jatropha curcas] |
| 17 |
Hb_027295_040 |
0.2149906674 |
- |
- |
hypothetical protein MIMGU_mgv1a017500mg [Erythranthe guttata] |
| 18 |
Hb_106415_010 |
0.2168708933 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X1 [Jatropha curcas] |
| 19 |
Hb_002084_010 |
0.217967129 |
- |
- |
hypothetical protein VITISV_017990 [Vitis vinifera] |
| 20 |
Hb_178620_010 |
0.217994038 |
- |
- |
ADP-ribosylation factor, arf, putative [Ricinus communis] |