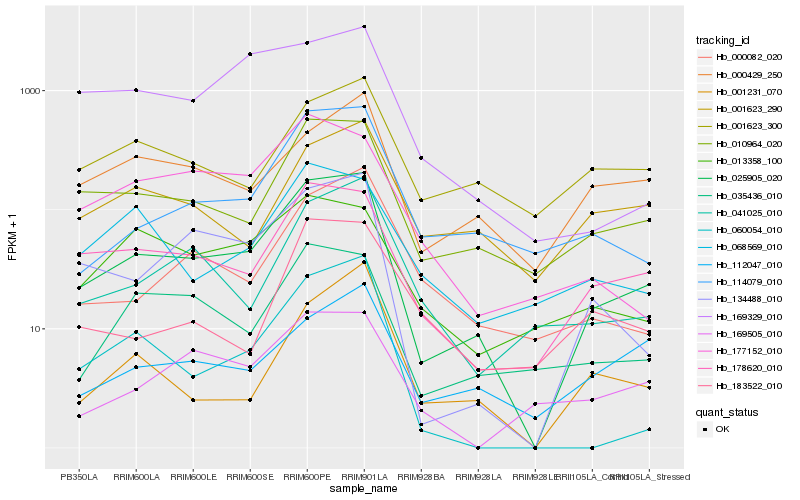
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025905_020 |
0.0 |
- |
- |
major allergen Pru ar 1-like [Jatropha curcas] |
| 2 |
Hb_010964_020 |
0.1499939082 |
- |
- |
RNA helicase [Arabidopsis thaliana] |
| 3 |
Hb_178620_010 |
0.1509081678 |
- |
- |
ADP-ribosylation factor, arf, putative [Ricinus communis] |
| 4 |
Hb_134488_010 |
0.157652047 |
- |
- |
- |
| 5 |
Hb_114079_010 |
0.1784574237 |
- |
- |
hypothetical protein CICLE_v10008147mg [Citrus clementina] |
| 6 |
Hb_035436_010 |
0.1785670188 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10-B-like [Populus euphratica] |
| 7 |
Hb_041025_010 |
0.1890003786 |
- |
- |
PREDICTED: uncharacterized protein LOC105635593 [Jatropha curcas] |
| 8 |
Hb_000082_020 |
0.1961672997 |
- |
- |
- |
| 9 |
Hb_000429_250 |
0.1975662749 |
- |
- |
hypothetical protein L484_000744 [Morus notabilis] |
| 10 |
Hb_060054_010 |
0.2003817475 |
- |
- |
NADH-ubiquinone oxidoreductase subunit B17.2, putative [Ricinus communis] |
| 11 |
Hb_169505_010 |
0.2033421387 |
- |
- |
hypothetical protein JCGZ_04480 [Jatropha curcas] |
| 12 |
Hb_177152_010 |
0.2056459669 |
- |
- |
glycosyl hydrolase family 38 family protein [Populus trichocarpa] |
| 13 |
Hb_013358_100 |
0.2059187789 |
- |
- |
unknown [Medicago truncatula] |
| 14 |
Hb_068569_010 |
0.2060280068 |
- |
- |
- |
| 15 |
Hb_001231_070 |
0.2062053341 |
- |
- |
- |
| 16 |
Hb_001623_300 |
0.2084123951 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13-B [Jatropha curcas] |
| 17 |
Hb_001623_290 |
0.2085941104 |
- |
- |
unknown [Populus trichocarpa] |
| 18 |
Hb_112047_010 |
0.2089528413 |
- |
- |
- |
| 19 |
Hb_169329_010 |
0.2153635045 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X2 [Jatropha curcas] |
| 20 |
Hb_183522_010 |
0.2188795242 |
- |
- |
Far upstream element-binding protein, putative [Ricinus communis] |