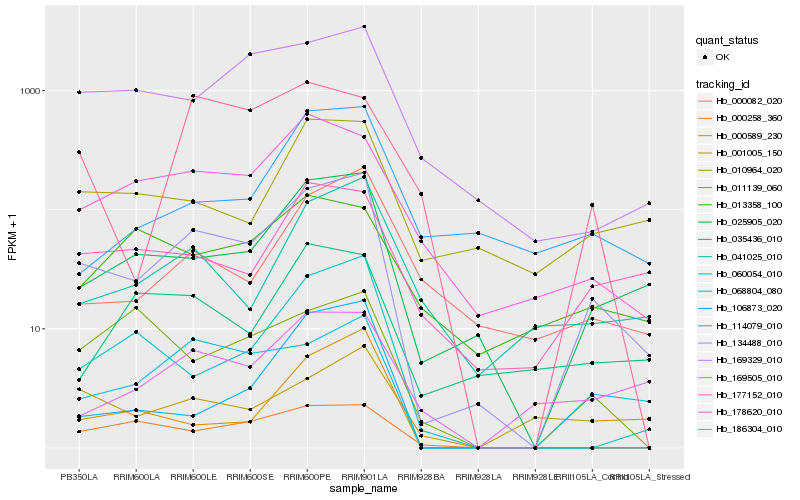
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_134488_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_025905_020 |
0.157652047 |
- |
- |
major allergen Pru ar 1-like [Jatropha curcas] |
| 3 |
Hb_068804_080 |
0.1779364679 |
- |
- |
PREDICTED: protein BRICK 1 [Jatropha curcas] |
| 4 |
Hb_177152_010 |
0.2092245428 |
- |
- |
glycosyl hydrolase family 38 family protein [Populus trichocarpa] |
| 5 |
Hb_169329_010 |
0.2138122898 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X2 [Jatropha curcas] |
| 6 |
Hb_041025_010 |
0.2161114549 |
- |
- |
PREDICTED: uncharacterized protein LOC105635593 [Jatropha curcas] |
| 7 |
Hb_178620_010 |
0.2176662152 |
- |
- |
ADP-ribosylation factor, arf, putative [Ricinus communis] |
| 8 |
Hb_000082_020 |
0.2230024998 |
- |
- |
- |
| 9 |
Hb_060054_010 |
0.2261037551 |
- |
- |
NADH-ubiquinone oxidoreductase subunit B17.2, putative [Ricinus communis] |
| 10 |
Hb_010964_020 |
0.2295293687 |
- |
- |
RNA helicase [Arabidopsis thaliana] |
| 11 |
Hb_169505_010 |
0.2310934616 |
- |
- |
hypothetical protein JCGZ_04480 [Jatropha curcas] |
| 12 |
Hb_000258_360 |
0.2407080249 |
- |
- |
PREDICTED: uncharacterized protein LOC104455487 [Eucalyptus grandis] |
| 13 |
Hb_001005_150 |
0.242791682 |
- |
- |
- |
| 14 |
Hb_114079_010 |
0.2437028638 |
- |
- |
hypothetical protein CICLE_v10008147mg [Citrus clementina] |
| 15 |
Hb_035436_010 |
0.245959638 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10-B-like [Populus euphratica] |
| 16 |
Hb_000589_230 |
0.2504959726 |
- |
- |
unknown [Picea sitchensis] |
| 17 |
Hb_013358_100 |
0.2542117841 |
- |
- |
unknown [Medicago truncatula] |
| 18 |
Hb_186304_010 |
0.255238111 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |
| 19 |
Hb_106873_020 |
0.2558399125 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 20 |
Hb_011139_060 |
0.2588393685 |
- |
- |
- |