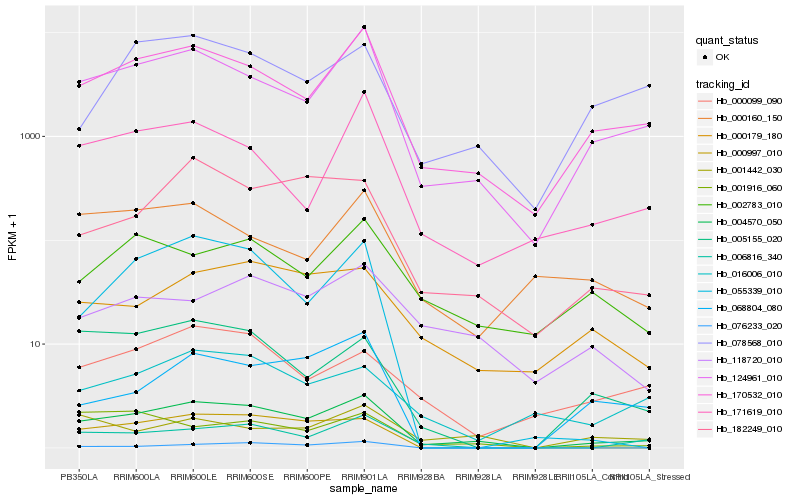
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004570_050 |
0.0 |
- |
- |
kinase, putative [Ricinus communis] |
| 2 |
Hb_170532_010 |
0.1288664342 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 8 [Jatropha curcas] |
| 3 |
Hb_124961_010 |
0.1366977694 |
- |
- |
hypothetical protein EUGRSUZ_B03893 [Eucalyptus grandis] |
| 4 |
Hb_006816_340 |
0.1820150325 |
- |
- |
- |
| 5 |
Hb_182249_010 |
0.1839541731 |
- |
- |
- |
| 6 |
Hb_000179_180 |
0.1933475131 |
- |
- |
hypothetical protein EUGRSUZ_C031461, partial [Eucalyptus grandis] |
| 7 |
Hb_005155_020 |
0.1974276884 |
- |
- |
- |
| 8 |
Hb_000997_010 |
0.1975958207 |
- |
- |
PREDICTED: FHA domain-containing protein At4g14490 isoform X2 [Jatropha curcas] |
| 9 |
Hb_171619_010 |
0.2061192123 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_001916_060 |
0.2078669739 |
- |
- |
PREDICTED: putative protein FAR1-RELATED SEQUENCE 10 isoform X2 [Jatropha curcas] |
| 11 |
Hb_002783_010 |
0.2081243876 |
- |
- |
PREDICTED: uncharacterized protein LOC105635318 [Jatropha curcas] |
| 12 |
Hb_016006_010 |
0.2103459821 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X2 [Jatropha curcas] |
| 13 |
Hb_118720_010 |
0.2114729912 |
- |
- |
PREDICTED: uncharacterized protein LOC105634948 [Jatropha curcas] |
| 14 |
Hb_055339_010 |
0.2121852211 |
- |
- |
EARLY-RESPONSIVE TO DEHYDRATION 7 family protein [Populus trichocarpa] |
| 15 |
Hb_078568_010 |
0.2122774508 |
- |
- |
hypothetical protein PRUPE_ppa013245mg [Prunus persica] |
| 16 |
Hb_000099_090 |
0.2154543535 |
- |
- |
Na(+)/H(+) antiporter, putative [Ricinus communis] |
| 17 |
Hb_076233_020 |
0.2158225501 |
transcription factor |
TF Family: C2H2 |
PREDICTED: dnaJ homolog subfamily C member 21 [Populus euphratica] |
| 18 |
Hb_000160_150 |
0.2176907825 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 3, chloroplastic [Cucumis melo] |
| 19 |
Hb_001442_030 |
0.2190959548 |
- |
- |
Peroxisome biogenesis factor [Medicago truncatula] |
| 20 |
Hb_068804_080 |
0.2195918474 |
- |
- |
PREDICTED: protein BRICK 1 [Jatropha curcas] |