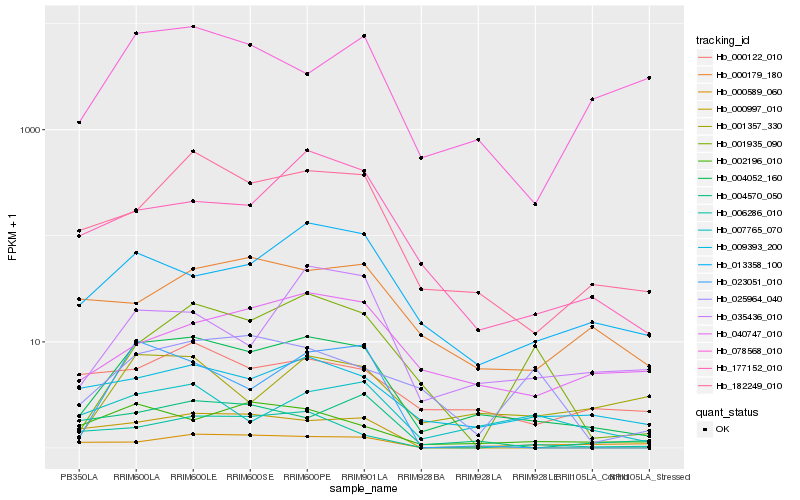
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004052_160 |
0.0 |
- |
- |
Chain A, Rna Binding Protein |
| 2 |
Hb_182249_010 |
0.1734128628 |
- |
- |
- |
| 3 |
Hb_009393_200 |
0.1997291795 |
- |
- |
PREDICTED: F-box only protein 13 [Jatropha curcas] |
| 4 |
Hb_013358_100 |
0.210766543 |
- |
- |
unknown [Medicago truncatula] |
| 5 |
Hb_002196_010 |
0.2211167461 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: proline-rich receptor-like protein kinase PERK1 [Jatropha curcas] |
| 6 |
Hb_040747_010 |
0.2234987164 |
- |
- |
hypothetical protein POPTR_0009s15120g [Populus trichocarpa] |
| 7 |
Hb_000122_010 |
0.227204923 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 8 |
Hb_177152_010 |
0.2275399714 |
- |
- |
glycosyl hydrolase family 38 family protein [Populus trichocarpa] |
| 9 |
Hb_001357_330 |
0.2278829978 |
- |
- |
PREDICTED: paraspeckle component 1 [Jatropha curcas] |
| 10 |
Hb_007765_070 |
0.2289895047 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 2 [Jatropha curcas] |
| 11 |
Hb_004570_050 |
0.2302113212 |
- |
- |
kinase, putative [Ricinus communis] |
| 12 |
Hb_001935_090 |
0.2348929835 |
- |
- |
PREDICTED: uncharacterized protein LOC105643180 isoform X1 [Jatropha curcas] |
| 13 |
Hb_023051_010 |
0.2367694854 |
- |
- |
PREDICTED: putative amidase C869.01 [Populus euphratica] |
| 14 |
Hb_035436_010 |
0.2381823981 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10-B-like [Populus euphratica] |
| 15 |
Hb_000179_180 |
0.2384949589 |
- |
- |
hypothetical protein EUGRSUZ_C031461, partial [Eucalyptus grandis] |
| 16 |
Hb_006286_010 |
0.2391083943 |
- |
- |
hypothetical protein PRUPE_ppa017148mg [Prunus persica] |
| 17 |
Hb_078568_010 |
0.2411117413 |
- |
- |
hypothetical protein PRUPE_ppa013245mg [Prunus persica] |
| 18 |
Hb_000589_060 |
0.2438973973 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_00699 [Jatropha curcas] |
| 19 |
Hb_025964_040 |
0.2446008344 |
- |
- |
PREDICTED: putative gamma-glutamylcyclotransferase At3g02910 [Jatropha curcas] |
| 20 |
Hb_000997_010 |
0.2478551298 |
- |
- |
PREDICTED: FHA domain-containing protein At4g14490 isoform X2 [Jatropha curcas] |