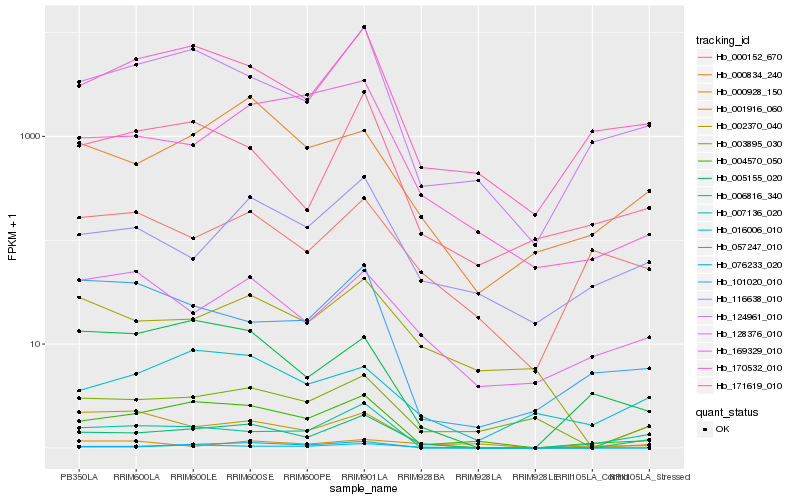
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006816_340 |
0.0 |
- |
- |
- |
| 2 |
Hb_004570_050 |
0.1820150325 |
- |
- |
kinase, putative [Ricinus communis] |
| 3 |
Hb_124961_010 |
0.1935069124 |
- |
- |
hypothetical protein EUGRSUZ_B03893 [Eucalyptus grandis] |
| 4 |
Hb_170532_010 |
0.1956683551 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 8 [Jatropha curcas] |
| 5 |
Hb_007136_020 |
0.1991100719 |
- |
- |
PREDICTED: oxygen-dependent coproporphyrinogen-III oxidase, chloroplastic [Populus euphratica] |
| 6 |
Hb_003895_030 |
0.2185136961 |
- |
- |
- |
| 7 |
Hb_171619_010 |
0.2195010532 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 8 |
Hb_000834_240 |
0.2219255452 |
- |
- |
hypothetical protein JCGZ_07230 [Jatropha curcas] |
| 9 |
Hb_116638_010 |
0.2267568464 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_057247_010 |
0.2346786681 |
- |
- |
hypothetical protein JCGZ_04800 [Jatropha curcas] |
| 11 |
Hb_128376_010 |
0.2357123059 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_076233_020 |
0.2395769756 |
transcription factor |
TF Family: C2H2 |
PREDICTED: dnaJ homolog subfamily C member 21 [Populus euphratica] |
| 13 |
Hb_016006_010 |
0.2404570037 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X2 [Jatropha curcas] |
| 14 |
Hb_000928_150 |
0.2416677635 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000152_670 |
0.2445782649 |
- |
- |
hypothetical protein VITISV_037603 [Vitis vinifera] |
| 16 |
Hb_002370_040 |
0.2454252402 |
- |
- |
PREDICTED: uncharacterized protein LOC105644336 [Jatropha curcas] |
| 17 |
Hb_005155_020 |
0.2467407708 |
- |
- |
- |
| 18 |
Hb_001916_060 |
0.2470723761 |
- |
- |
PREDICTED: putative protein FAR1-RELATED SEQUENCE 10 isoform X2 [Jatropha curcas] |
| 19 |
Hb_101020_010 |
0.2493001756 |
- |
- |
- |
| 20 |
Hb_169329_010 |
0.2499897577 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X2 [Jatropha curcas] |