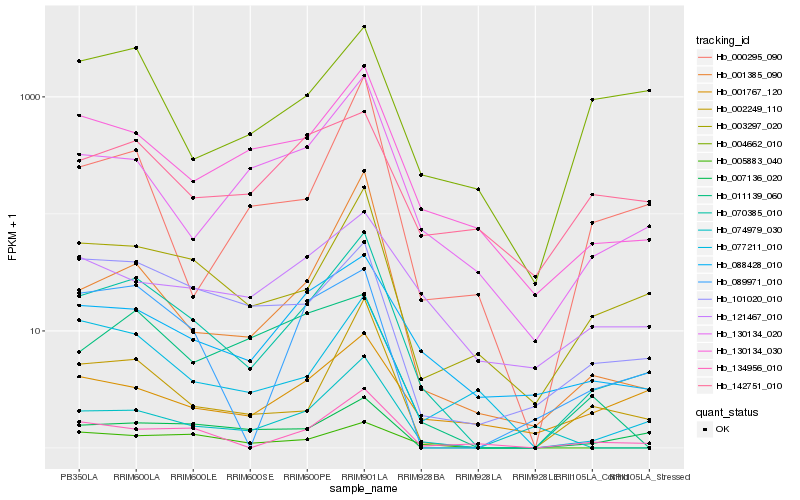
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_070385_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103323653 [Prunus mume] |
| 2 |
Hb_003297_020 |
0.1659776811 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 3 |
Hb_002249_110 |
0.1759282499 |
- |
- |
PREDICTED: centromere protein S isoform X1 [Jatropha curcas] |
| 4 |
Hb_130134_030 |
0.1857273426 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_14472 [Jatropha curcas] |
| 5 |
Hb_134956_010 |
0.1881599498 |
- |
- |
PREDICTED: (S)-coclaurine N-methyltransferase [Jatropha curcas] |
| 6 |
Hb_088428_010 |
0.1913817624 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181 homolog [Jatropha curcas] |
| 7 |
Hb_130134_020 |
0.2042020258 |
- |
- |
- |
| 8 |
Hb_101020_010 |
0.2052044454 |
- |
- |
- |
| 9 |
Hb_007136_020 |
0.2089295969 |
- |
- |
PREDICTED: oxygen-dependent coproporphyrinogen-III oxidase, chloroplastic [Populus euphratica] |
| 10 |
Hb_089971_010 |
0.209129132 |
- |
- |
PREDICTED: (S)-coclaurine N-methyltransferase [Cucumis sativus] |
| 11 |
Hb_077211_010 |
0.2099514782 |
- |
- |
PREDICTED: lariat debranching enzyme-like [Jatropha curcas] |
| 12 |
Hb_005883_040 |
0.2184819102 |
- |
- |
hypothetical protein JCGZ_11546 [Jatropha curcas] |
| 13 |
Hb_001385_090 |
0.2212681977 |
- |
- |
PREDICTED: uncharacterized protein LOC105644404 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000295_090 |
0.2291623103 |
- |
- |
Clathrin light chain 2 [Glycine soja] |
| 15 |
Hb_074979_030 |
0.234435318 |
- |
- |
HSP20-like chaperones superfamily protein [Theobroma cacao] |
| 16 |
Hb_121467_010 |
0.2350077491 |
- |
- |
tubulin-specific chaperone E, putative [Ricinus communis] |
| 17 |
Hb_001767_120 |
0.2411466266 |
- |
- |
- |
| 18 |
Hb_142751_010 |
0.2447266339 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_011139_060 |
0.2481766617 |
- |
- |
- |
| 20 |
Hb_004662_010 |
0.2493053988 |
- |
- |
Nascent polypeptide-associated complex subunit beta [Medicago truncatula] |