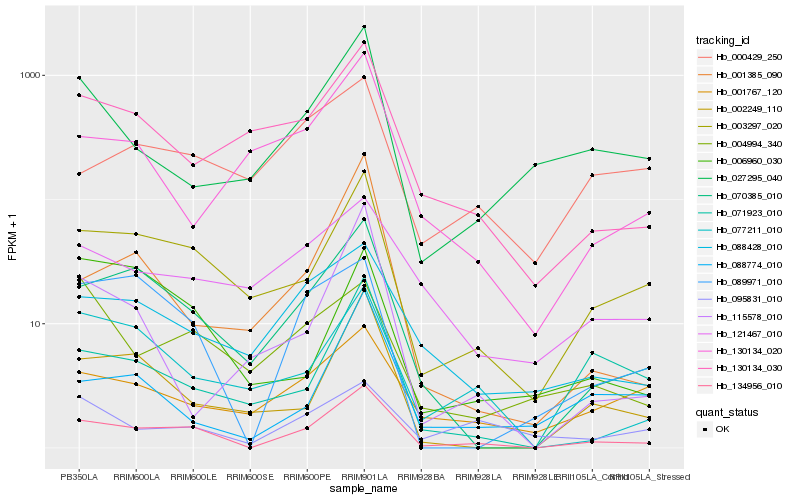
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_134956_010 |
0.0 |
- |
- |
PREDICTED: (S)-coclaurine N-methyltransferase [Jatropha curcas] |
| 2 |
Hb_003297_020 |
0.1837383315 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 3 |
Hb_070385_010 |
0.1881599498 |
- |
- |
PREDICTED: uncharacterized protein LOC103323653 [Prunus mume] |
| 4 |
Hb_002249_110 |
0.232319529 |
- |
- |
PREDICTED: centromere protein S isoform X1 [Jatropha curcas] |
| 5 |
Hb_088428_010 |
0.237464177 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181 homolog [Jatropha curcas] |
| 6 |
Hb_089971_010 |
0.2384091568 |
- |
- |
PREDICTED: (S)-coclaurine N-methyltransferase [Cucumis sativus] |
| 7 |
Hb_077211_010 |
0.2387205514 |
- |
- |
PREDICTED: lariat debranching enzyme-like [Jatropha curcas] |
| 8 |
Hb_130134_030 |
0.2444289898 |
transcription factor |
TF Family: C3H |
hypothetical protein JCGZ_14472 [Jatropha curcas] |
| 9 |
Hb_071923_010 |
0.2458087549 |
- |
- |
- |
| 10 |
Hb_001767_120 |
0.2529617333 |
- |
- |
- |
| 11 |
Hb_027295_040 |
0.2536288644 |
- |
- |
hypothetical protein MIMGU_mgv1a017500mg [Erythranthe guttata] |
| 12 |
Hb_088774_010 |
0.2549990587 |
- |
- |
- |
| 13 |
Hb_001385_090 |
0.2587267121 |
- |
- |
PREDICTED: uncharacterized protein LOC105644404 isoform X2 [Jatropha curcas] |
| 14 |
Hb_121467_010 |
0.2650562674 |
- |
- |
tubulin-specific chaperone E, putative [Ricinus communis] |
| 15 |
Hb_115578_010 |
0.2680499089 |
- |
- |
PREDICTED: prefoldin subunit 1-like [Populus euphratica] |
| 16 |
Hb_095831_010 |
0.2683113282 |
- |
- |
PREDICTED: uncharacterized protein LOC104884649 [Beta vulgaris subsp. vulgaris] |
| 17 |
Hb_130134_020 |
0.270174854 |
- |
- |
- |
| 18 |
Hb_000429_250 |
0.2749952311 |
- |
- |
hypothetical protein L484_000744 [Morus notabilis] |
| 19 |
Hb_006960_030 |
0.2764718286 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB44-like [Jatropha curcas] |
| 20 |
Hb_004994_340 |
0.2771524199 |
- |
- |
- |