| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
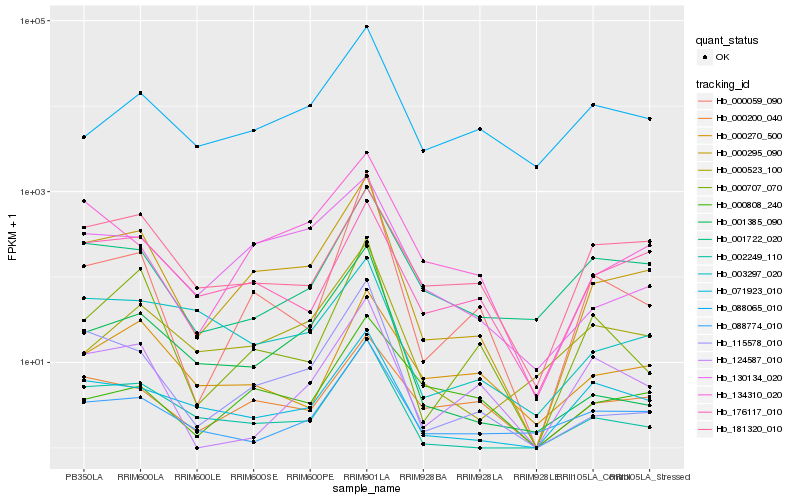
Hb_000295_090 |
0.0 |
- |
- |
Clathrin light chain 2 [Glycine soja] |
| 2 |
Hb_002249_110 |
0.143982607 |
- |
- |
PREDICTED: centromere protein S isoform X1 [Jatropha curcas] |
| 3 |
Hb_115578_010 |
0.1565610238 |
- |
- |
PREDICTED: prefoldin subunit 1-like [Populus euphratica] |
| 4 |
Hb_130134_020 |
0.1766368033 |
- |
- |
- |
| 5 |
Hb_088774_010 |
0.1767418252 |
- |
- |
- |
| 6 |
Hb_001385_090 |
0.181064029 |
- |
- |
PREDICTED: uncharacterized protein LOC105644404 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000523_100 |
0.1827973026 |
- |
- |
PREDICTED: protein DJ-1 homolog D-like [Populus euphratica] |
| 8 |
Hb_001722_020 |
0.183036524 |
- |
- |
- |
| 9 |
Hb_071923_010 |
0.1846470244 |
- |
- |
- |
| 10 |
Hb_134310_020 |
0.1847251876 |
- |
- |
PREDICTED: putative cytochrome c oxidase subunit 5C-4-like [Citrus sinensis] |
| 11 |
Hb_000059_090 |
0.1883073948 |
- |
- |
- |
| 12 |
Hb_000707_070 |
0.2009954745 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.6-like isoform X1 [Populus euphratica] |
| 13 |
Hb_000200_040 |
0.2041025759 |
- |
- |
- |
| 14 |
Hb_088065_010 |
0.2041614719 |
- |
- |
hypothetical protein glysoja_007934 [Glycine soja] |
| 15 |
Hb_176117_010 |
0.2068581785 |
- |
- |
Cold-inducible RNA-binding protein [Morus notabilis] |
| 16 |
Hb_124587_010 |
0.2071994485 |
- |
- |
PREDICTED: importin-5-like [Musa acuminata subsp. malaccensis] |
| 17 |
Hb_000270_500 |
0.2163095808 |
- |
- |
PREDICTED: uncharacterized protein LOC105640439 [Jatropha curcas] |
| 18 |
Hb_003297_020 |
0.2224155508 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 19 |
Hb_000808_240 |
0.2251430995 |
- |
- |
- |
| 20 |
Hb_181320_010 |
0.2255359656 |
- |
- |
PREDICTED: signal recognition particle 14 kDa protein [Jatropha curcas] |