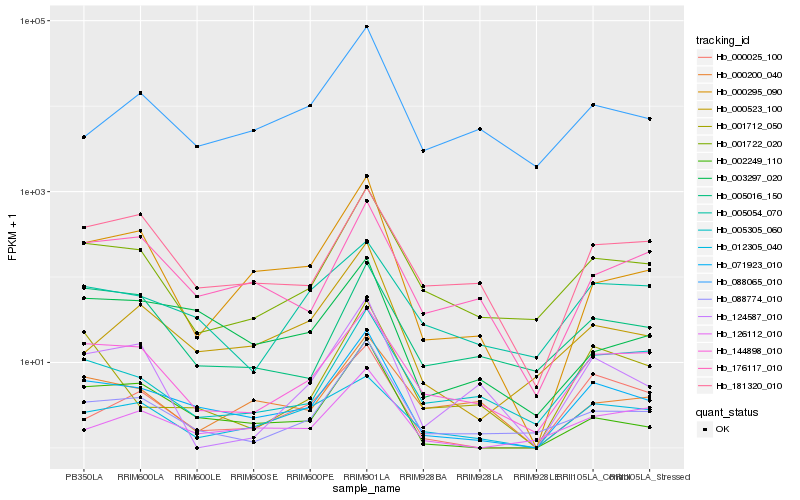
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_071923_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_001722_020 |
0.1637470175 |
- |
- |
- |
| 3 |
Hb_002249_110 |
0.1644235314 |
- |
- |
PREDICTED: centromere protein S isoform X1 [Jatropha curcas] |
| 4 |
Hb_088774_010 |
0.1729444255 |
- |
- |
- |
| 5 |
Hb_144898_010 |
0.1799212423 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |
| 6 |
Hb_000295_090 |
0.1846470244 |
- |
- |
Clathrin light chain 2 [Glycine soja] |
| 7 |
Hb_003297_020 |
0.1874798217 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 8 |
Hb_176117_010 |
0.1932851267 |
- |
- |
Cold-inducible RNA-binding protein [Morus notabilis] |
| 9 |
Hb_181320_010 |
0.1938198819 |
- |
- |
PREDICTED: signal recognition particle 14 kDa protein [Jatropha curcas] |
| 10 |
Hb_126112_010 |
0.1943862911 |
- |
- |
PREDICTED: uncharacterized protein LOC105636715 [Jatropha curcas] |
| 11 |
Hb_000523_100 |
0.1973633641 |
- |
- |
PREDICTED: protein DJ-1 homolog D-like [Populus euphratica] |
| 12 |
Hb_000025_100 |
0.199407972 |
- |
- |
- |
| 13 |
Hb_005305_060 |
0.2009432102 |
- |
- |
hypothetical protein MIMGU_mgv1a015111mg [Erythranthe guttata] |
| 14 |
Hb_005054_070 |
0.2068192014 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_001712_050 |
0.2117543875 |
- |
- |
PREDICTED: uncharacterized protein LOC105641944 [Jatropha curcas] |
| 16 |
Hb_088065_010 |
0.2151400864 |
- |
- |
hypothetical protein glysoja_007934 [Glycine soja] |
| 17 |
Hb_012305_040 |
0.2162005344 |
- |
- |
PREDICTED: DNA-directed RNA polymerases II, IV and V subunit 9A-like [Jatropha curcas] |
| 18 |
Hb_000200_040 |
0.216482578 |
- |
- |
- |
| 19 |
Hb_005016_150 |
0.2209767616 |
- |
- |
- |
| 20 |
Hb_124587_010 |
0.2213308895 |
- |
- |
PREDICTED: importin-5-like [Musa acuminata subsp. malaccensis] |