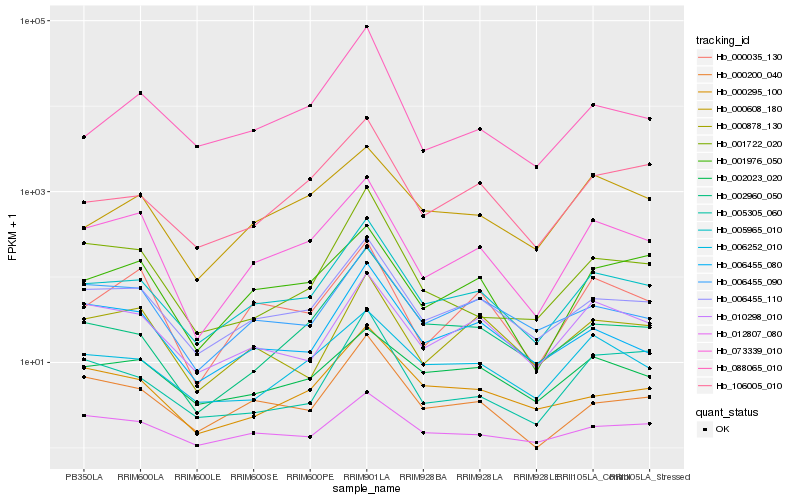
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005965_010 |
0.0 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb family transcription factor APL-like [Jatropha curcas] |
| 2 |
Hb_006455_110 |
0.0730914773 |
- |
- |
bromodomain-containing protein, putative [Ricinus communis] |
| 3 |
Hb_073339_010 |
0.120301266 |
- |
- |
hypothetical protein 30 [Hevea brasiliensis] |
| 4 |
Hb_106005_010 |
0.1298829371 |
- |
- |
EG5651 [Manihot esculenta] |
| 5 |
Hb_012807_080 |
0.1342711994 |
- |
- |
hypothetical protein CICLE_v10028099mg [Citrus clementina] |
| 6 |
Hb_006455_080 |
0.1363532952 |
- |
- |
PREDICTED: transcription factor GTE6-like isoform X2 [Nelumbo nucifera] |
| 7 |
Hb_006455_090 |
0.1379021411 |
- |
- |
PREDICTED: transcription factor GTE1-like [Jatropha curcas] |
| 8 |
Hb_002960_050 |
0.1394517168 |
- |
- |
PREDICTED: uncharacterized protein LOC105635325 [Jatropha curcas] |
| 9 |
Hb_000295_100 |
0.1409121654 |
- |
- |
- |
| 10 |
Hb_000200_040 |
0.1419568114 |
- |
- |
- |
| 11 |
Hb_000608_180 |
0.144537734 |
- |
- |
PREDICTED: putative 4-hydroxy-4-methyl-2-oxoglutarate aldolase 2 [Tarenaya hassleriana] |
| 12 |
Hb_006252_010 |
0.1446117855 |
- |
- |
PREDICTED: alpha-ketoglutarate-dependent dioxygenase alkB homolog 6 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001976_050 |
0.1525137119 |
- |
- |
PREDICTED: protein SKIP34 [Jatropha curcas] |
| 14 |
Hb_010298_010 |
0.1532894384 |
- |
- |
PREDICTED: uncharacterized protein At2g02148 [Jatropha curcas] |
| 15 |
Hb_000878_130 |
0.1551990392 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005305_060 |
0.1558083859 |
- |
- |
hypothetical protein MIMGU_mgv1a015111mg [Erythranthe guttata] |
| 17 |
Hb_000035_130 |
0.15674829 |
- |
- |
hypothetical protein Csa_4G652025 [Cucumis sativus] |
| 18 |
Hb_088065_010 |
0.1575136156 |
- |
- |
hypothetical protein glysoja_007934 [Glycine soja] |
| 19 |
Hb_002023_020 |
0.1594534539 |
- |
- |
PREDICTED: aladin isoform X2 [Jatropha curcas] |
| 20 |
Hb_001722_020 |
0.1595181475 |
- |
- |
- |