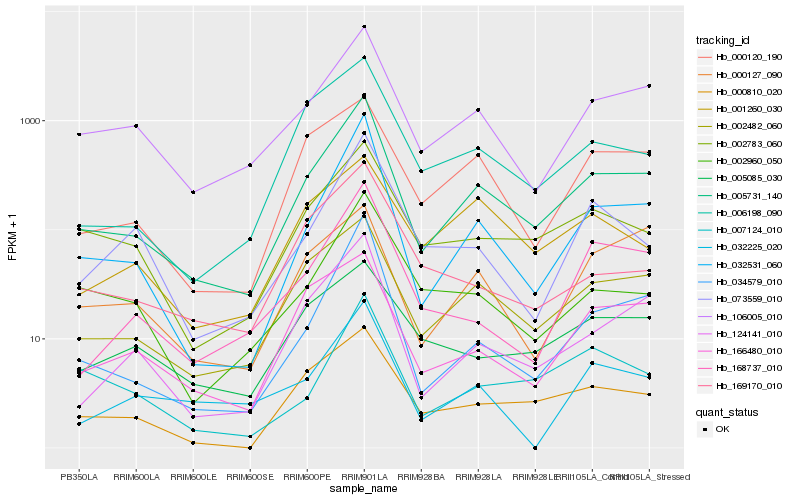
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005731_140 |
0.0 |
- |
- |
hypothetical protein JCGZ_26808 [Jatropha curcas] |
| 2 |
Hb_032531_060 |
0.1145780929 |
- |
- |
hypothetical protein PRUPE_ppa017035mg, partial [Prunus persica] |
| 3 |
Hb_002482_060 |
0.1161127987 |
- |
- |
hypothetical protein CICLE_v10033884mg, partial [Citrus clementina] |
| 4 |
Hb_106005_010 |
0.1251189792 |
- |
- |
EG5651 [Manihot esculenta] |
| 5 |
Hb_124141_010 |
0.1340539327 |
- |
- |
plant origin recognition complex subunit, putative [Ricinus communis] |
| 6 |
Hb_034579_010 |
0.1431126302 |
- |
- |
PREDICTED: nudix hydrolase 27, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000810_020 |
0.1453194038 |
- |
- |
- |
| 8 |
Hb_002783_060 |
0.1508723553 |
- |
- |
PREDICTED: uncharacterized protein LOC105635325 [Jatropha curcas] |
| 9 |
Hb_006198_090 |
0.1572593462 |
- |
- |
PREDICTED: vesicle-associated membrane protein 721-like isoform X2 [Tarenaya hassleriana] |
| 10 |
Hb_000120_190 |
0.1580006456 |
- |
- |
Pre-mRNA-splicing factor ini1 [Ricinus communis] |
| 11 |
Hb_166480_010 |
0.1631216563 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase, mitochondrial-like [Cucumis melo] |
| 12 |
Hb_168737_010 |
0.1693786289 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_002960_050 |
0.1708457891 |
- |
- |
PREDICTED: uncharacterized protein LOC105635325 [Jatropha curcas] |
| 14 |
Hb_007124_010 |
0.1743043647 |
- |
- |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_073559_010 |
0.1771213875 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 12 [Jatropha curcas] |
| 16 |
Hb_169170_010 |
0.1802781472 |
- |
- |
PREDICTED: protein disulfide-isomerase 5-1 isoform X2 [Nelumbo nucifera] |
| 17 |
Hb_005085_030 |
0.1853201104 |
- |
- |
PREDICTED: N-acetyltransferase 9-like protein isoform X1 [Jatropha curcas] |
| 18 |
Hb_032225_020 |
0.1884666388 |
- |
- |
hypothetical protein POPTR_0006s08140g [Populus trichocarpa] |
| 19 |
Hb_001260_030 |
0.1887968616 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000127_090 |
0.1897813106 |
- |
- |
conserved hypothetical protein [Ricinus communis] |