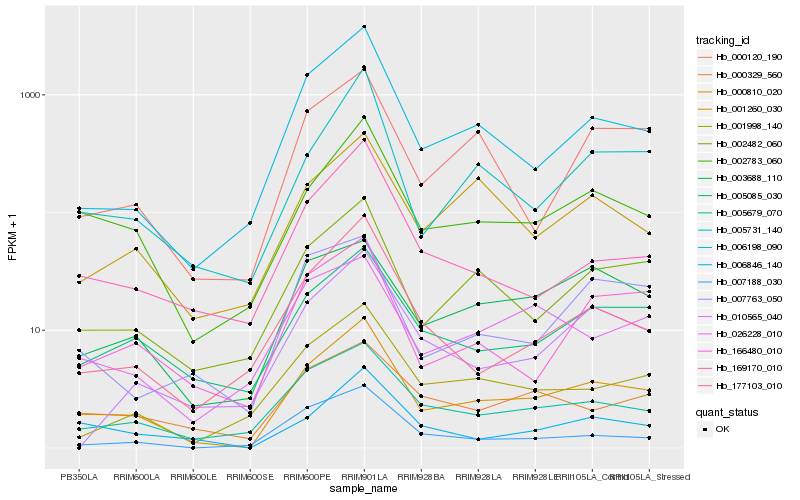
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000810_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_002783_060 |
0.1173942424 |
- |
- |
PREDICTED: uncharacterized protein LOC105635325 [Jatropha curcas] |
| 3 |
Hb_002482_060 |
0.1390586805 |
- |
- |
hypothetical protein CICLE_v10033884mg, partial [Citrus clementina] |
| 4 |
Hb_006198_090 |
0.1416597258 |
- |
- |
PREDICTED: vesicle-associated membrane protein 721-like isoform X2 [Tarenaya hassleriana] |
| 5 |
Hb_005679_070 |
0.1435571512 |
- |
- |
hypothetical protein M569_07741, partial [Genlisea aurea] |
| 6 |
Hb_005731_140 |
0.1453194038 |
- |
- |
hypothetical protein JCGZ_26808 [Jatropha curcas] |
| 7 |
Hb_005085_030 |
0.146651924 |
- |
- |
PREDICTED: N-acetyltransferase 9-like protein isoform X1 [Jatropha curcas] |
| 8 |
Hb_006846_140 |
0.1555649157 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_166480_010 |
0.1625336989 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase, mitochondrial-like [Cucumis melo] |
| 10 |
Hb_000120_190 |
0.1655589914 |
- |
- |
Pre-mRNA-splicing factor ini1 [Ricinus communis] |
| 11 |
Hb_001998_140 |
0.1684673436 |
- |
- |
- |
| 12 |
Hb_003688_110 |
0.1688369974 |
- |
- |
- |
| 13 |
Hb_177103_010 |
0.1708563014 |
- |
- |
PREDICTED: histone H2AX [Elaeis guineensis] |
| 14 |
Hb_000329_560 |
0.1736170948 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001260_030 |
0.1751159036 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_026228_010 |
0.1753975856 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_169170_010 |
0.1760014888 |
- |
- |
PREDICTED: protein disulfide-isomerase 5-1 isoform X2 [Nelumbo nucifera] |
| 18 |
Hb_007763_050 |
0.1792167775 |
- |
- |
hypothetical protein L484_012752 [Morus notabilis] |
| 19 |
Hb_007188_030 |
0.1817087894 |
- |
- |
PREDICTED: DNA topoisomerase 3-alpha-like [Malus domestica] |
| 20 |
Hb_010565_040 |
0.1820280381 |
- |
- |
hypothetical protein POPTR_0015s05280g, partial [Populus trichocarpa] |