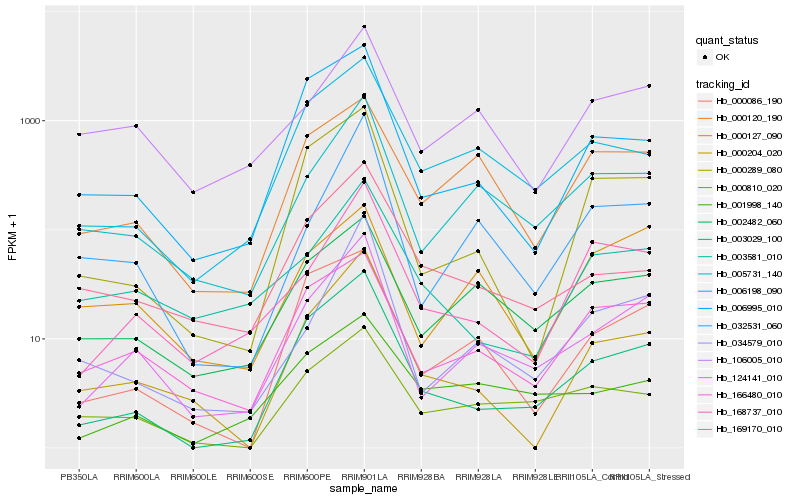
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_124141_010 |
0.0 |
- |
- |
plant origin recognition complex subunit, putative [Ricinus communis] |
| 2 |
Hb_005731_140 |
0.1340539327 |
- |
- |
hypothetical protein JCGZ_26808 [Jatropha curcas] |
| 3 |
Hb_003029_100 |
0.1564227096 |
- |
- |
- |
| 4 |
Hb_166480_010 |
0.1616378337 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase, mitochondrial-like [Cucumis melo] |
| 5 |
Hb_032531_060 |
0.1617147702 |
- |
- |
hypothetical protein PRUPE_ppa017035mg, partial [Prunus persica] |
| 6 |
Hb_034579_010 |
0.1630047827 |
- |
- |
PREDICTED: nudix hydrolase 27, chloroplastic [Jatropha curcas] |
| 7 |
Hb_002482_060 |
0.167198769 |
- |
- |
hypothetical protein CICLE_v10033884mg, partial [Citrus clementina] |
| 8 |
Hb_168737_010 |
0.177989076 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_106005_010 |
0.1787278095 |
- |
- |
EG5651 [Manihot esculenta] |
| 10 |
Hb_006198_090 |
0.182655125 |
- |
- |
PREDICTED: vesicle-associated membrane protein 721-like isoform X2 [Tarenaya hassleriana] |
| 11 |
Hb_000204_020 |
0.1829752206 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized N-acetyltransferase ycf52 [Jatropha curcas] |
| 12 |
Hb_000086_190 |
0.1842805217 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 3-like protein [Cucumis sativus] |
| 13 |
Hb_000289_080 |
0.1859282586 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_006995_010 |
0.1906403132 |
- |
- |
PREDICTED: 14 kDa zinc-binding protein [Jatropha curcas] |
| 15 |
Hb_000810_020 |
0.1914851729 |
- |
- |
- |
| 16 |
Hb_000120_190 |
0.1985536998 |
- |
- |
Pre-mRNA-splicing factor ini1 [Ricinus communis] |
| 17 |
Hb_001998_140 |
0.199863782 |
- |
- |
- |
| 18 |
Hb_169170_010 |
0.201688424 |
- |
- |
PREDICTED: protein disulfide-isomerase 5-1 isoform X2 [Nelumbo nucifera] |
| 19 |
Hb_000127_090 |
0.2064206755 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003581_010 |
0.2107420639 |
- |
- |
hypothetical protein VITISV_004687 [Vitis vinifera] |