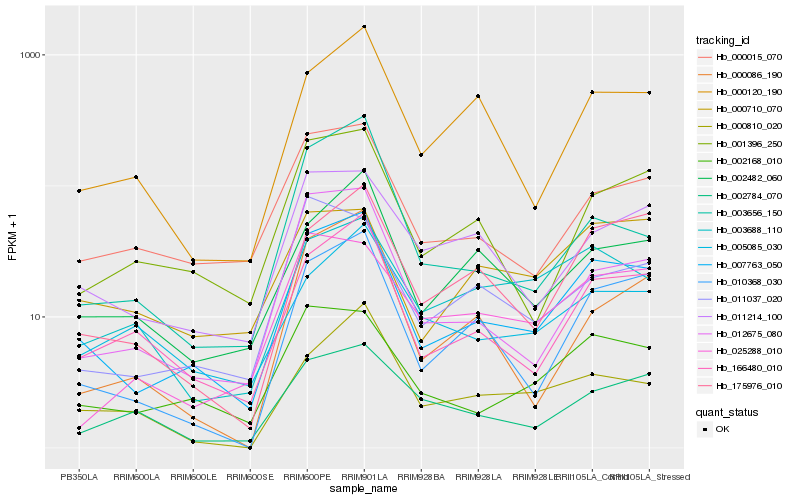
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007763_050 |
0.0 |
- |
- |
hypothetical protein L484_012752 [Morus notabilis] |
| 2 |
Hb_166480_010 |
0.1270883384 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase, mitochondrial-like [Cucumis melo] |
| 3 |
Hb_002168_010 |
0.1336335395 |
- |
- |
PREDICTED: bifunctional dihydrofolate reductase-thymidylate synthase-like isoform X4 [Vitis vinifera] |
| 4 |
Hb_010368_030 |
0.1343189716 |
- |
- |
BnaA07g35570D [Brassica napus] |
| 5 |
Hb_000015_070 |
0.1411237648 |
- |
- |
hypothetical protein POPTR_0001s22690g [Populus trichocarpa] |
| 6 |
Hb_001396_250 |
0.1435612869 |
- |
- |
PREDICTED: stress-associated endoplasmic reticulum protein 2 [Jatropha curcas] |
| 7 |
Hb_175976_010 |
0.1490299271 |
- |
- |
- |
| 8 |
Hb_012675_080 |
0.1522171791 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 1 [Populus euphratica] |
| 9 |
Hb_002482_060 |
0.1534783693 |
- |
- |
hypothetical protein CICLE_v10033884mg, partial [Citrus clementina] |
| 10 |
Hb_011214_100 |
0.1544251722 |
- |
- |
PREDICTED: uncharacterized protein LOC105633003 [Jatropha curcas] |
| 11 |
Hb_000120_190 |
0.160669628 |
- |
- |
Pre-mRNA-splicing factor ini1 [Ricinus communis] |
| 12 |
Hb_011037_020 |
0.1712435561 |
- |
- |
unknown [Picea sitchensis] |
| 13 |
Hb_003688_110 |
0.1729893187 |
- |
- |
- |
| 14 |
Hb_000710_070 |
0.1747410007 |
- |
- |
PREDICTED: uncharacterized protein LOC105645553 [Jatropha curcas] |
| 15 |
Hb_025288_010 |
0.1759158258 |
- |
- |
PREDICTED: protein unc-50 homolog [Jatropha curcas] |
| 16 |
Hb_000810_020 |
0.1792167775 |
- |
- |
- |
| 17 |
Hb_002784_070 |
0.179253391 |
- |
- |
BnaUnng01470D [Brassica napus] |
| 18 |
Hb_005085_030 |
0.1799343022 |
- |
- |
PREDICTED: N-acetyltransferase 9-like protein isoform X1 [Jatropha curcas] |
| 19 |
Hb_003656_150 |
0.1817637858 |
- |
- |
- |
| 20 |
Hb_000086_190 |
0.1835567543 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 3-like protein [Cucumis sativus] |