| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
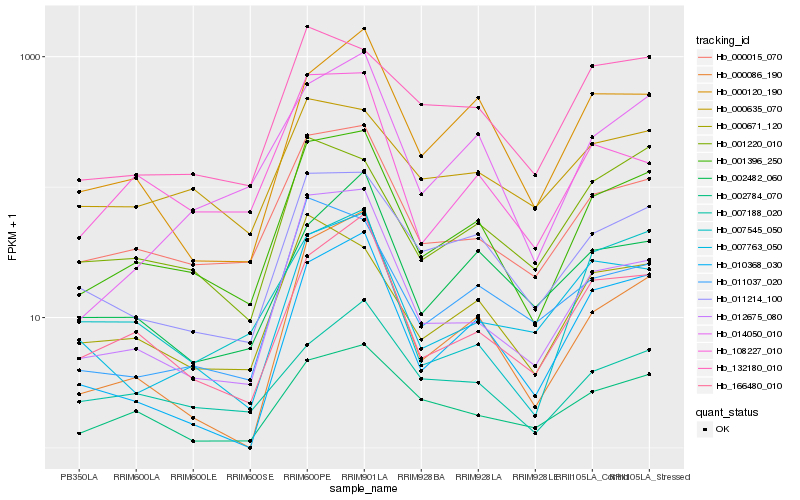
Hb_001396_250 |
0.0 |
- |
- |
PREDICTED: stress-associated endoplasmic reticulum protein 2 [Jatropha curcas] |
| 2 |
Hb_000015_070 |
0.0865252283 |
- |
- |
hypothetical protein POPTR_0001s22690g [Populus trichocarpa] |
| 3 |
Hb_011214_100 |
0.1131961311 |
- |
- |
PREDICTED: uncharacterized protein LOC105633003 [Jatropha curcas] |
| 4 |
Hb_166480_010 |
0.1198548704 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase, mitochondrial-like [Cucumis melo] |
| 5 |
Hb_012675_080 |
0.1289622674 |
- |
- |
PREDICTED: cytoplasmic tRNA 2-thiolation protein 1 [Populus euphratica] |
| 6 |
Hb_014050_010 |
0.1318904208 |
- |
- |
PREDICTED: histone H3.3-like, partial [Phoenix dactylifera] |
| 7 |
Hb_002784_070 |
0.1371619504 |
- |
- |
BnaUnng01470D [Brassica napus] |
| 8 |
Hb_000671_120 |
0.1401049155 |
- |
- |
hypothetical protein JCGZ_23777 [Jatropha curcas] |
| 9 |
Hb_007763_050 |
0.1435612869 |
- |
- |
hypothetical protein L484_012752 [Morus notabilis] |
| 10 |
Hb_010368_030 |
0.1461237441 |
- |
- |
BnaA07g35570D [Brassica napus] |
| 11 |
Hb_108227_010 |
0.1477460147 |
- |
- |
RecName: Full=Profilin-2; AltName: Full=Pollen allergen Hev b 8.0102; AltName: Allergen=Hev b 8.0102 [Hevea brasiliensis] |
| 12 |
Hb_000120_190 |
0.1498839118 |
- |
- |
Pre-mRNA-splicing factor ini1 [Ricinus communis] |
| 13 |
Hb_132180_010 |
0.1520063989 |
- |
- |
PREDICTED: protein SPIRAL1-like 1 [Populus euphratica] |
| 14 |
Hb_007188_020 |
0.1522969499 |
- |
- |
hypothetical protein PRUPE_ppb010436mg [Prunus persica] |
| 15 |
Hb_011037_020 |
0.1526695898 |
- |
- |
unknown [Picea sitchensis] |
| 16 |
Hb_000086_190 |
0.1533969294 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 3-like protein [Cucumis sativus] |
| 17 |
Hb_001220_010 |
0.1559964649 |
- |
- |
hypothetical protein JCGZ_00411 [Jatropha curcas] |
| 18 |
Hb_000635_070 |
0.1594814129 |
- |
- |
PREDICTED: probable signal peptidase complex subunit 1 [Jatropha curcas] |
| 19 |
Hb_007545_050 |
0.1606574246 |
- |
- |
- |
| 20 |
Hb_002482_060 |
0.1621806712 |
- |
- |
hypothetical protein CICLE_v10033884mg, partial [Citrus clementina] |