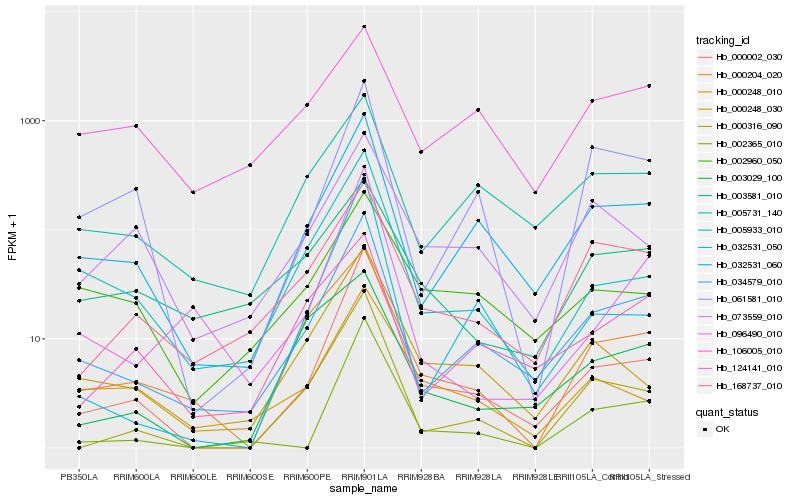
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_034579_010 |
0.0 |
- |
- |
PREDICTED: nudix hydrolase 27, chloroplastic [Jatropha curcas] |
| 2 |
Hb_032531_060 |
0.0779168568 |
- |
- |
hypothetical protein PRUPE_ppa017035mg, partial [Prunus persica] |
| 3 |
Hb_005731_140 |
0.1431126302 |
- |
- |
hypothetical protein JCGZ_26808 [Jatropha curcas] |
| 4 |
Hb_124141_010 |
0.1630047827 |
- |
- |
plant origin recognition complex subunit, putative [Ricinus communis] |
| 5 |
Hb_000002_030 |
0.164444902 |
- |
- |
- |
| 6 |
Hb_005933_010 |
0.1707897168 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Populus euphratica] |
| 7 |
Hb_168737_010 |
0.1766073702 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_000316_090 |
0.1863706845 |
- |
- |
- |
| 9 |
Hb_000204_020 |
0.186500904 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized N-acetyltransferase ycf52 [Jatropha curcas] |
| 10 |
Hb_061581_010 |
0.1876930492 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_000248_030 |
0.1879996162 |
- |
- |
hypothetical protein Pmar_PMAR005331 [Perkinsus marinus ATCC 50983] |
| 12 |
Hb_032531_050 |
0.2013753648 |
- |
- |
hypothetical protein CICLE_v10003980mg [Citrus clementina] |
| 13 |
Hb_096490_010 |
0.202125514 |
- |
- |
hypothetical protein CICLE_v100210772mg, partial [Citrus clementina] |
| 14 |
Hb_106005_010 |
0.2095516548 |
- |
- |
EG5651 [Manihot esculenta] |
| 15 |
Hb_073559_010 |
0.2137758875 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 12 [Jatropha curcas] |
| 16 |
Hb_002960_050 |
0.2166145541 |
- |
- |
PREDICTED: uncharacterized protein LOC105635325 [Jatropha curcas] |
| 17 |
Hb_003029_100 |
0.2179994952 |
- |
- |
- |
| 18 |
Hb_000248_010 |
0.2202642338 |
- |
- |
hypothetical protein PRUPE_ppa008056mg [Prunus persica] |
| 19 |
Hb_003581_010 |
0.2220465801 |
- |
- |
hypothetical protein VITISV_004687 [Vitis vinifera] |
| 20 |
Hb_002365_010 |
0.224649182 |
- |
- |
hypothetical protein CISIN_1g0405342mg, partial [Citrus sinensis] |