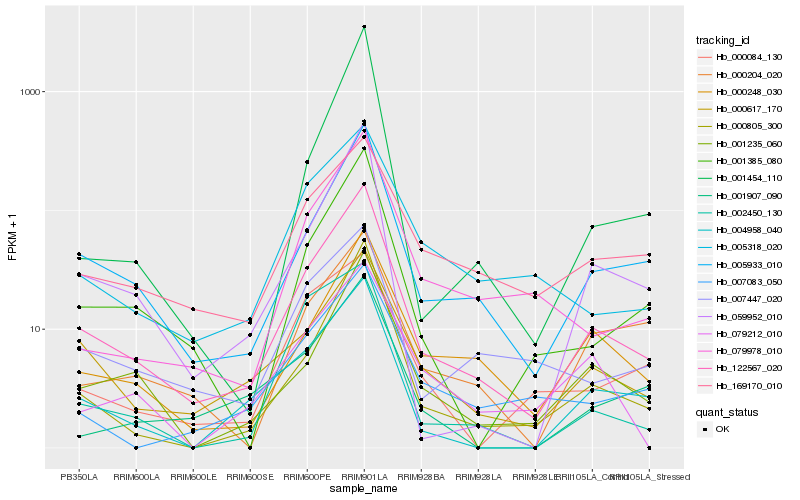
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_122567_020 |
0.0 |
- |
- |
hypothetical protein CICLE_v10009900mg [Citrus clementina] |
| 2 |
Hb_005933_010 |
0.1066502484 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Populus euphratica] |
| 3 |
Hb_000805_300 |
0.1324449682 |
- |
- |
- |
| 4 |
Hb_000617_170 |
0.1472916569 |
- |
- |
- |
| 5 |
Hb_059952_010 |
0.1478638433 |
- |
- |
PREDICTED: probable protein S-acyltransferase 7 [Eucalyptus grandis] |
| 6 |
Hb_001385_080 |
0.1498491892 |
- |
- |
- |
| 7 |
Hb_001235_060 |
0.1509662296 |
- |
- |
- |
| 8 |
Hb_000248_030 |
0.1512647604 |
- |
- |
hypothetical protein Pmar_PMAR005331 [Perkinsus marinus ATCC 50983] |
| 9 |
Hb_004958_040 |
0.1545817694 |
- |
- |
- |
| 10 |
Hb_079978_010 |
0.1677401923 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP71 isoform X1 [Nelumbo nucifera] |
| 11 |
Hb_005318_020 |
0.1722976343 |
- |
- |
PREDICTED: lecithin-cholesterol acyltransferase-like 4 isoform X3 [Jatropha curcas] |
| 12 |
Hb_007447_020 |
0.1752838217 |
- |
- |
hypothetical protein CISIN_1g038065mg [Citrus sinensis] |
| 13 |
Hb_007083_050 |
0.180881398 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 12-like isoform X2 [Nelumbo nucifera] |
| 14 |
Hb_169170_010 |
0.1871155555 |
- |
- |
PREDICTED: protein disulfide-isomerase 5-1 isoform X2 [Nelumbo nucifera] |
| 15 |
Hb_001907_090 |
0.1902932644 |
- |
- |
- |
| 16 |
Hb_079212_010 |
0.1942878005 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000204_020 |
0.1947297701 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized N-acetyltransferase ycf52 [Jatropha curcas] |
| 18 |
Hb_000084_130 |
0.196645958 |
- |
- |
- |
| 19 |
Hb_001454_110 |
0.1975842328 |
- |
- |
hypothetical protein POPTR_0026s00250g [Populus trichocarpa] |
| 20 |
Hb_002450_130 |
0.1985277922 |
- |
- |
hypothetical protein JCGZ_06565 [Jatropha curcas] |