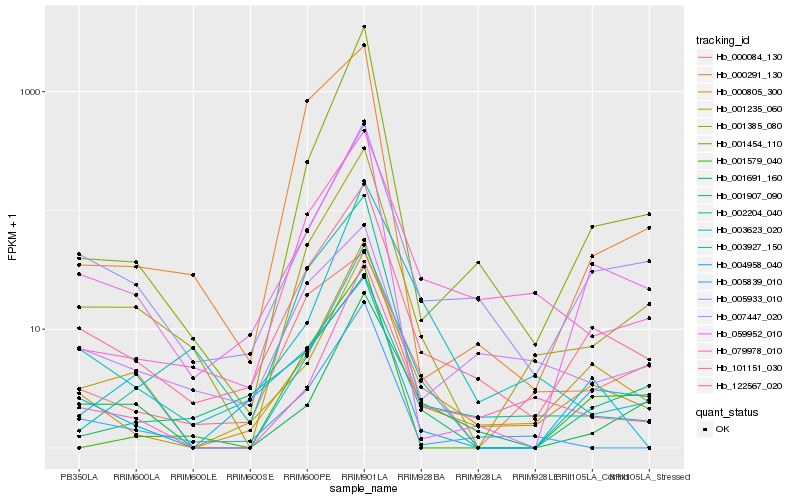
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001385_080 |
0.0 |
- |
- |
- |
| 2 |
Hb_122567_020 |
0.1498491892 |
- |
- |
hypothetical protein CICLE_v10009900mg [Citrus clementina] |
| 3 |
Hb_005933_010 |
0.1613796237 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Populus euphratica] |
| 4 |
Hb_059952_010 |
0.1716866252 |
- |
- |
PREDICTED: probable protein S-acyltransferase 7 [Eucalyptus grandis] |
| 5 |
Hb_000805_300 |
0.1770458949 |
- |
- |
- |
| 6 |
Hb_001454_110 |
0.1859151464 |
- |
- |
hypothetical protein POPTR_0026s00250g [Populus trichocarpa] |
| 7 |
Hb_101151_030 |
0.1893623231 |
- |
- |
- |
| 8 |
Hb_079978_010 |
0.1898489953 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP71 isoform X1 [Nelumbo nucifera] |
| 9 |
Hb_001235_060 |
0.1963463873 |
- |
- |
- |
| 10 |
Hb_005839_010 |
0.199292393 |
- |
- |
30S ribosomal protein s13 [Camellia sinensis] |
| 11 |
Hb_000291_130 |
0.2011768621 |
- |
- |
- |
| 12 |
Hb_001691_160 |
0.2013853001 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002204_040 |
0.2021190929 |
- |
- |
hypothetical protein B456_008G181200 [Gossypium raimondii] |
| 14 |
Hb_000084_130 |
0.2040515016 |
- |
- |
- |
| 15 |
Hb_001907_090 |
0.208269271 |
- |
- |
- |
| 16 |
Hb_001579_040 |
0.2112010001 |
- |
- |
- |
| 17 |
Hb_007447_020 |
0.2112025139 |
- |
- |
hypothetical protein CISIN_1g038065mg [Citrus sinensis] |
| 18 |
Hb_003927_150 |
0.2139815799 |
- |
- |
- |
| 19 |
Hb_004958_040 |
0.2183159841 |
- |
- |
- |
| 20 |
Hb_003623_020 |
0.2209959672 |
- |
- |
- |