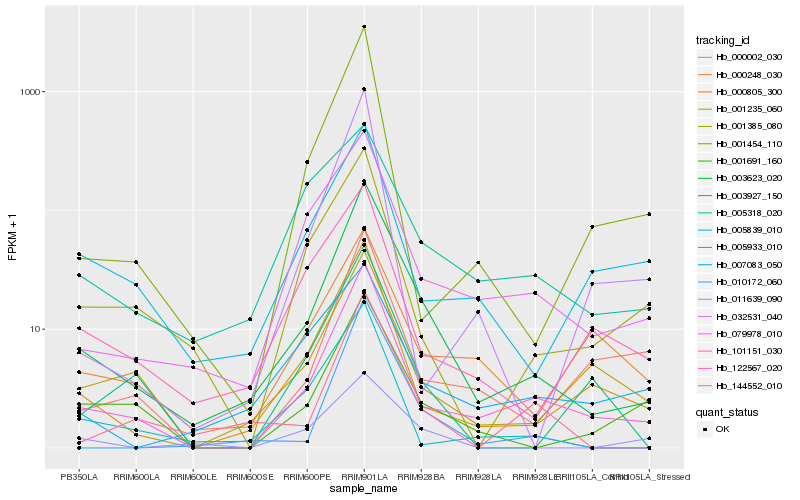
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003623_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_101151_030 |
0.1562425755 |
- |
- |
- |
| 3 |
Hb_000805_300 |
0.1758974756 |
- |
- |
- |
| 4 |
Hb_079978_010 |
0.1934994735 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP71 isoform X1 [Nelumbo nucifera] |
| 5 |
Hb_010172_060 |
0.1992837315 |
- |
- |
PREDICTED: uncharacterized protein LOC105634266 [Jatropha curcas] |
| 6 |
Hb_144552_010 |
0.2042374834 |
- |
- |
CC-NBS-LRR protein, putative [Theobroma cacao] |
| 7 |
Hb_001691_160 |
0.2169473812 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_011639_090 |
0.2173694483 |
- |
- |
PREDICTED: transcription factor bHLH75-like [Populus euphratica] |
| 9 |
Hb_005839_010 |
0.2175290408 |
- |
- |
30S ribosomal protein s13 [Camellia sinensis] |
| 10 |
Hb_001235_060 |
0.2200823043 |
- |
- |
- |
| 11 |
Hb_001385_080 |
0.2209959672 |
- |
- |
- |
| 12 |
Hb_122567_020 |
0.2216383911 |
- |
- |
hypothetical protein CICLE_v10009900mg [Citrus clementina] |
| 13 |
Hb_001454_110 |
0.223692506 |
- |
- |
hypothetical protein POPTR_0026s00250g [Populus trichocarpa] |
| 14 |
Hb_003927_150 |
0.2246076756 |
- |
- |
- |
| 15 |
Hb_005933_010 |
0.2363294714 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Populus euphratica] |
| 16 |
Hb_000002_030 |
0.2379795711 |
- |
- |
- |
| 17 |
Hb_005318_020 |
0.2381304139 |
- |
- |
PREDICTED: lecithin-cholesterol acyltransferase-like 4 isoform X3 [Jatropha curcas] |
| 18 |
Hb_007083_050 |
0.2436265974 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 12-like isoform X2 [Nelumbo nucifera] |
| 19 |
Hb_000248_030 |
0.2456944042 |
- |
- |
hypothetical protein Pmar_PMAR005331 [Perkinsus marinus ATCC 50983] |
| 20 |
Hb_032531_040 |
0.2458512243 |
- |
- |
E3 ISG15--protein ligase HERC5 [Gossypium arboreum] |