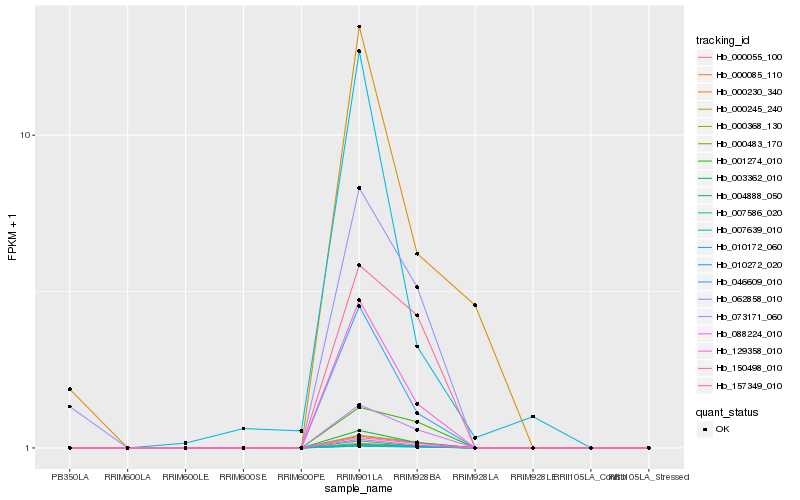
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_129358_010 |
0.0 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 11-like [Jatropha curcas] |
| 2 |
Hb_007639_010 |
0.0266312071 |
- |
- |
hypothetical protein JCGZ_11284 [Jatropha curcas] |
| 3 |
Hb_046609_010 |
0.0307627206 |
- |
- |
- |
| 4 |
Hb_003362_010 |
0.0614040525 |
- |
- |
Actin-related protein 2/3 complex subunit 4 [Morus notabilis] |
| 5 |
Hb_150498_010 |
0.076843558 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 6 |
Hb_010272_020 |
0.1050877987 |
- |
- |
hypothetical protein VITISV_030841 [Vitis vinifera] |
| 7 |
Hb_073171_060 |
0.1166543974 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 8 |
Hb_000055_100 |
0.1293795951 |
- |
- |
polyprotein [Oryza australiensis] |
| 9 |
Hb_000368_130 |
0.1301430033 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_088224_010 |
0.1348134252 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 11 |
Hb_000245_240 |
0.1360447783 |
- |
- |
- |
| 12 |
Hb_000085_110 |
0.1369726166 |
- |
- |
hypothetical protein JCGZ_00957 [Jatropha curcas] |
| 13 |
Hb_004888_050 |
0.1734030533 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 14 |
Hb_007586_020 |
0.1749888604 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 15 |
Hb_062858_010 |
0.1897845568 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 16 |
Hb_010172_060 |
0.1928769169 |
- |
- |
PREDICTED: uncharacterized protein LOC105634266 [Jatropha curcas] |
| 17 |
Hb_157349_010 |
0.1987822161 |
- |
- |
PREDICTED: uncharacterized protein LOC102588546 [Solanum tuberosum] |
| 18 |
Hb_001274_010 |
0.2075208969 |
- |
- |
- |
| 19 |
Hb_000230_340 |
0.2184820621 |
- |
- |
Transcription elongation factor, putative [Ricinus communis] |
| 20 |
Hb_000483_170 |
0.2312872165 |
transcription factor |
TF Family: NAC |
PREDICTED: protein FEZ [Jatropha curcas] |