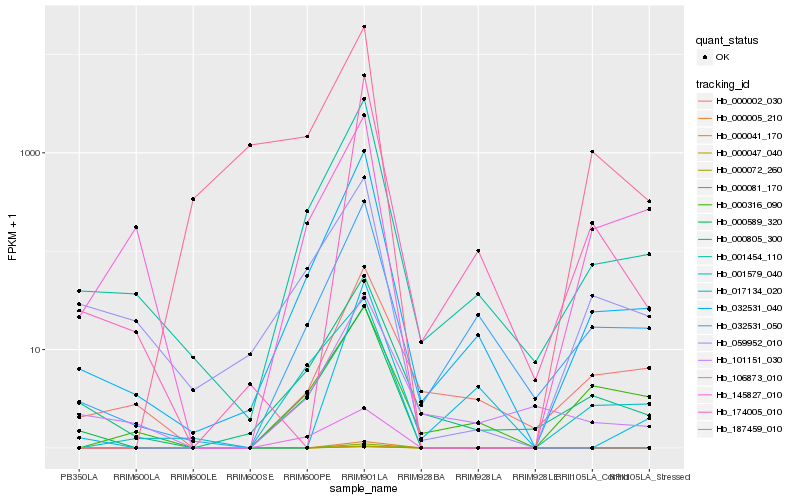
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032531_040 |
0.0 |
- |
- |
E3 ISG15--protein ligase HERC5 [Gossypium arboreum] |
| 2 |
Hb_001454_110 |
0.0693422252 |
- |
- |
hypothetical protein POPTR_0026s00250g [Populus trichocarpa] |
| 3 |
Hb_032531_050 |
0.1450428715 |
- |
- |
hypothetical protein CICLE_v10003980mg [Citrus clementina] |
| 4 |
Hb_000805_300 |
0.1549191652 |
- |
- |
- |
| 5 |
Hb_174005_010 |
0.1748062971 |
- |
- |
hypothetical protein EUGRSUZ_D00590 [Eucalyptus grandis] |
| 6 |
Hb_000589_320 |
0.1851923981 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_001579_040 |
0.1946877346 |
- |
- |
- |
| 8 |
Hb_059952_010 |
0.2013251611 |
- |
- |
PREDICTED: probable protein S-acyltransferase 7 [Eucalyptus grandis] |
| 9 |
Hb_000002_030 |
0.2024653166 |
- |
- |
- |
| 10 |
Hb_101151_030 |
0.2066898123 |
- |
- |
- |
| 11 |
Hb_000316_090 |
0.2085586777 |
- |
- |
- |
| 12 |
Hb_187459_010 |
0.2089232008 |
- |
- |
PREDICTED: uncharacterized protein LOC102619199, partial [Citrus sinensis] |
| 13 |
Hb_017134_020 |
0.2212982129 |
- |
- |
- |
| 14 |
Hb_145827_010 |
0.2306108415 |
- |
- |
hypothetical protein POPTR_0003s13630g [Populus trichocarpa] |
| 15 |
Hb_106873_010 |
0.2380677011 |
- |
- |
- |
| 16 |
Hb_000005_210 |
0.2382774326 |
- |
- |
- |
| 17 |
Hb_000041_170 |
0.2382774326 |
- |
- |
PREDICTED: uncharacterized protein At4g02000-like [Eucalyptus grandis] |
| 18 |
Hb_000047_040 |
0.2382774326 |
- |
- |
PREDICTED: uncharacterized protein LOC105775709 [Gossypium raimondii] |
| 19 |
Hb_000072_260 |
0.2382774326 |
- |
- |
hypothetical protein MTR_5g044090 [Medicago truncatula] |
| 20 |
Hb_000081_170 |
0.2382774326 |
- |
- |
- |