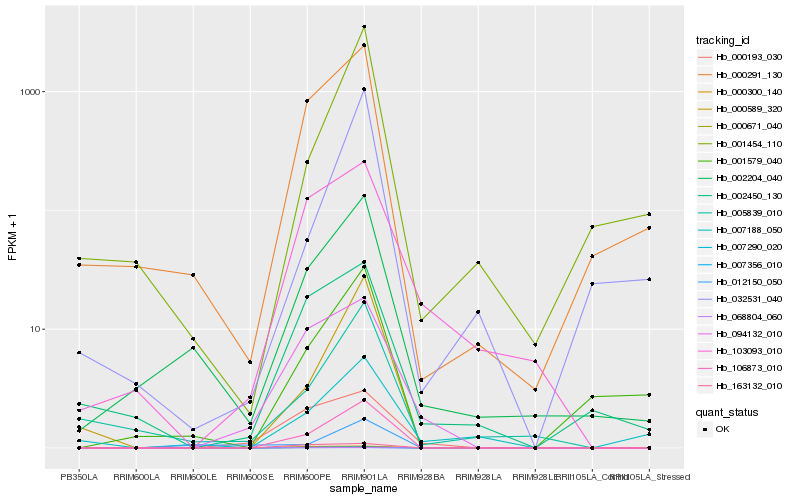
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_106873_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_000671_040 |
0.1078759866 |
- |
- |
- |
| 3 |
Hb_000589_320 |
0.1436802179 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_002204_040 |
0.20107034 |
- |
- |
hypothetical protein B456_008G181200 [Gossypium raimondii] |
| 5 |
Hb_000291_130 |
0.2013243832 |
- |
- |
- |
| 6 |
Hb_001579_040 |
0.221125234 |
- |
- |
- |
| 7 |
Hb_094132_010 |
0.2313536006 |
- |
- |
hypothetical protein POPTR_0018s03360g [Populus trichocarpa] |
| 8 |
Hb_001454_110 |
0.2323790741 |
- |
- |
hypothetical protein POPTR_0026s00250g [Populus trichocarpa] |
| 9 |
Hb_005839_010 |
0.2342228297 |
- |
- |
30S ribosomal protein s13 [Camellia sinensis] |
| 10 |
Hb_163132_010 |
0.2373651788 |
- |
- |
- |
| 11 |
Hb_032531_040 |
0.2380677011 |
- |
- |
E3 ISG15--protein ligase HERC5 [Gossypium arboreum] |
| 12 |
Hb_007356_010 |
0.2414689319 |
- |
- |
CTV.19 [Citrus trifoliata] |
| 13 |
Hb_007290_020 |
0.246642137 |
- |
- |
PREDICTED: subtilisin-like protease SBT4.14 [Jatropha curcas] |
| 14 |
Hb_068804_060 |
0.2467858563 |
- |
- |
PREDICTED: ABC transporter G family member 17-like [Jatropha curcas] |
| 15 |
Hb_103093_010 |
0.2506528106 |
- |
- |
PREDICTED: wall-associated receptor kinase 4-like [Elaeis guineensis] |
| 16 |
Hb_000300_140 |
0.2524121101 |
transcription factor |
TF Family: HB |
Protein WUSCHEL, putative [Ricinus communis] |
| 17 |
Hb_000193_030 |
0.2560559755 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Jatropha curcas] |
| 18 |
Hb_002450_130 |
0.2653377637 |
- |
- |
hypothetical protein JCGZ_06565 [Jatropha curcas] |
| 19 |
Hb_007188_050 |
0.2655982911 |
- |
- |
- |
| 20 |
Hb_012150_050 |
0.271511063 |
- |
- |
protein binding protein, putative [Ricinus communis] |