| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
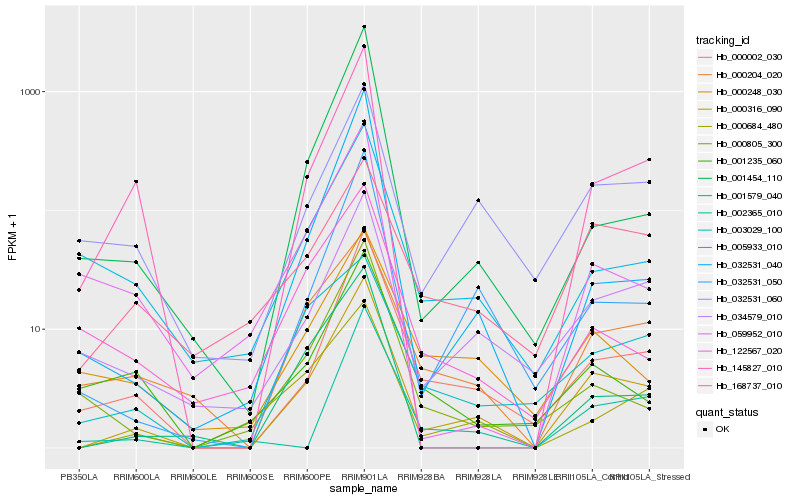
Hb_000316_090 |
0.0 |
- |
- |
- |
| 2 |
Hb_000002_030 |
0.1544650175 |
- |
- |
- |
| 3 |
Hb_032531_050 |
0.167814294 |
- |
- |
hypothetical protein CICLE_v10003980mg [Citrus clementina] |
| 4 |
Hb_034579_010 |
0.1863706845 |
- |
- |
PREDICTED: nudix hydrolase 27, chloroplastic [Jatropha curcas] |
| 5 |
Hb_145827_010 |
0.1877989993 |
- |
- |
hypothetical protein POPTR_0003s13630g [Populus trichocarpa] |
| 6 |
Hb_000684_480 |
0.1918966808 |
transcription factor |
TF Family: Orphans |
- |
| 7 |
Hb_001579_040 |
0.1942184949 |
- |
- |
- |
| 8 |
Hb_032531_060 |
0.1972647604 |
- |
- |
hypothetical protein PRUPE_ppa017035mg, partial [Prunus persica] |
| 9 |
Hb_000204_020 |
0.1980477769 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized N-acetyltransferase ycf52 [Jatropha curcas] |
| 10 |
Hb_001454_110 |
0.2048154726 |
- |
- |
hypothetical protein POPTR_0026s00250g [Populus trichocarpa] |
| 11 |
Hb_032531_040 |
0.2085586777 |
- |
- |
E3 ISG15--protein ligase HERC5 [Gossypium arboreum] |
| 12 |
Hb_000248_030 |
0.2101209233 |
- |
- |
hypothetical protein Pmar_PMAR005331 [Perkinsus marinus ATCC 50983] |
| 13 |
Hb_000805_300 |
0.2157965949 |
- |
- |
- |
| 14 |
Hb_005933_010 |
0.2194967953 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Populus euphratica] |
| 15 |
Hb_002365_010 |
0.2211442778 |
- |
- |
hypothetical protein CISIN_1g0405342mg, partial [Citrus sinensis] |
| 16 |
Hb_001235_060 |
0.229227557 |
- |
- |
- |
| 17 |
Hb_059952_010 |
0.2297451583 |
- |
- |
PREDICTED: probable protein S-acyltransferase 7 [Eucalyptus grandis] |
| 18 |
Hb_168737_010 |
0.2302942937 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_003029_100 |
0.2318484884 |
- |
- |
- |
| 20 |
Hb_122567_020 |
0.2348791581 |
- |
- |
hypothetical protein CICLE_v10009900mg [Citrus clementina] |