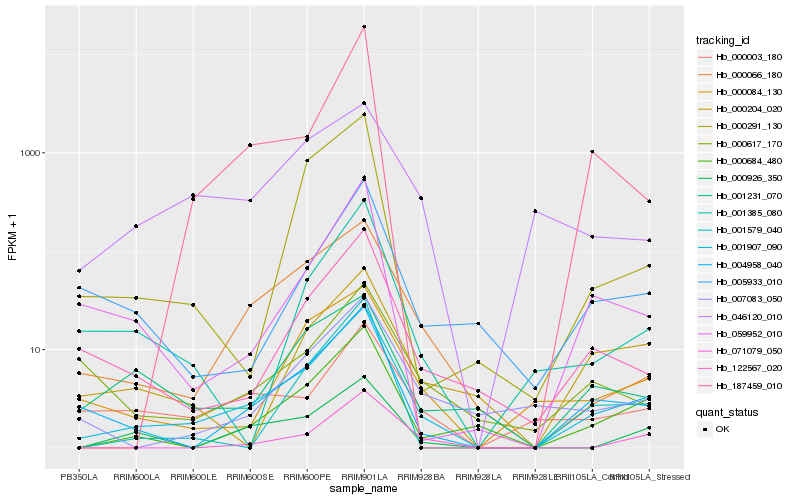
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001907_090 |
0.0 |
- |
- |
- |
| 2 |
Hb_004958_040 |
0.162008491 |
- |
- |
- |
| 3 |
Hb_000684_480 |
0.1817863765 |
transcription factor |
TF Family: Orphans |
- |
| 4 |
Hb_122567_020 |
0.1902932644 |
- |
- |
hypothetical protein CICLE_v10009900mg [Citrus clementina] |
| 5 |
Hb_000066_180 |
0.198334255 |
- |
- |
hypothetical protein POPTR_0008s02710g [Populus trichocarpa] |
| 6 |
Hb_001385_080 |
0.208269271 |
- |
- |
- |
| 7 |
Hb_000084_130 |
0.2114540729 |
- |
- |
- |
| 8 |
Hb_059952_010 |
0.211657962 |
- |
- |
PREDICTED: probable protein S-acyltransferase 7 [Eucalyptus grandis] |
| 9 |
Hb_007083_050 |
0.213596811 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 12-like isoform X2 [Nelumbo nucifera] |
| 10 |
Hb_001579_040 |
0.214637693 |
- |
- |
- |
| 11 |
Hb_000926_350 |
0.2193109111 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: upstream activation factor subunit UAF30 [Jatropha curcas] |
| 12 |
Hb_000204_020 |
0.2211457424 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized N-acetyltransferase ycf52 [Jatropha curcas] |
| 13 |
Hb_000003_180 |
0.2213357533 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_071079_050 |
0.2213378628 |
- |
- |
- |
| 15 |
Hb_005933_010 |
0.2213573031 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Populus euphratica] |
| 16 |
Hb_187459_010 |
0.2253612527 |
- |
- |
PREDICTED: uncharacterized protein LOC102619199, partial [Citrus sinensis] |
| 17 |
Hb_001231_070 |
0.2282432583 |
- |
- |
- |
| 18 |
Hb_000291_130 |
0.2309024979 |
- |
- |
- |
| 19 |
Hb_000617_170 |
0.2320402825 |
- |
- |
- |
| 20 |
Hb_046120_010 |
0.234230658 |
- |
- |
- |