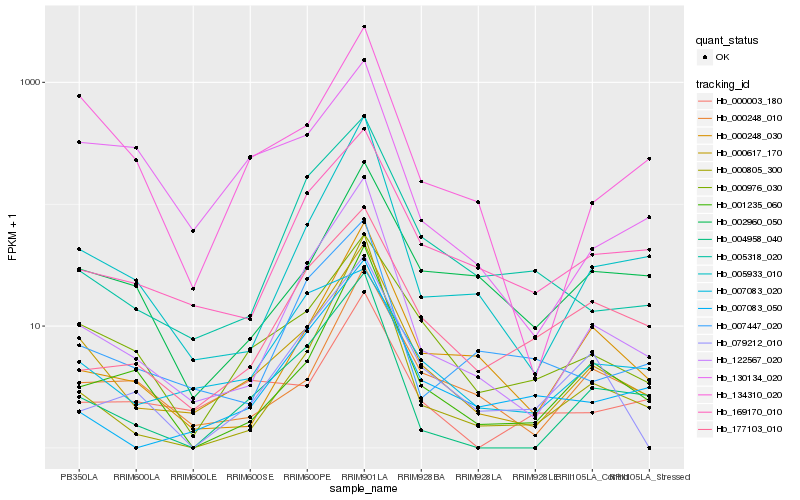
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000617_170 |
0.0 |
- |
- |
- |
| 2 |
Hb_122567_020 |
0.1472916569 |
- |
- |
hypothetical protein CICLE_v10009900mg [Citrus clementina] |
| 3 |
Hb_000976_030 |
0.1494227177 |
- |
- |
PREDICTED: molybdenum cofactor sulfurase-like [Citrus sinensis] |
| 4 |
Hb_005933_010 |
0.1622913935 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Populus euphratica] |
| 5 |
Hb_134310_020 |
0.1670042492 |
- |
- |
PREDICTED: putative cytochrome c oxidase subunit 5C-4-like [Citrus sinensis] |
| 6 |
Hb_000248_010 |
0.1738330666 |
- |
- |
hypothetical protein PRUPE_ppa008056mg [Prunus persica] |
| 7 |
Hb_169170_010 |
0.1791226011 |
- |
- |
PREDICTED: protein disulfide-isomerase 5-1 isoform X2 [Nelumbo nucifera] |
| 8 |
Hb_000248_030 |
0.1869258173 |
- |
- |
hypothetical protein Pmar_PMAR005331 [Perkinsus marinus ATCC 50983] |
| 9 |
Hb_004958_040 |
0.1931443871 |
- |
- |
- |
| 10 |
Hb_001235_060 |
0.1955732602 |
- |
- |
- |
| 11 |
Hb_005318_020 |
0.1966238373 |
- |
- |
PREDICTED: lecithin-cholesterol acyltransferase-like 4 isoform X3 [Jatropha curcas] |
| 12 |
Hb_002960_050 |
0.2027949293 |
- |
- |
PREDICTED: uncharacterized protein LOC105635325 [Jatropha curcas] |
| 13 |
Hb_007083_050 |
0.2053208178 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 12-like isoform X2 [Nelumbo nucifera] |
| 14 |
Hb_000003_180 |
0.2076515812 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_177103_010 |
0.2101903158 |
- |
- |
PREDICTED: histone H2AX [Elaeis guineensis] |
| 16 |
Hb_007083_020 |
0.2115409994 |
- |
- |
hypothetical protein F775_15403 [Aegilops tauschii] |
| 17 |
Hb_007447_020 |
0.2135989373 |
- |
- |
hypothetical protein CISIN_1g038065mg [Citrus sinensis] |
| 18 |
Hb_130134_020 |
0.2161611656 |
- |
- |
- |
| 19 |
Hb_079212_010 |
0.2192650121 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000805_300 |
0.2203698496 |
- |
- |
- |