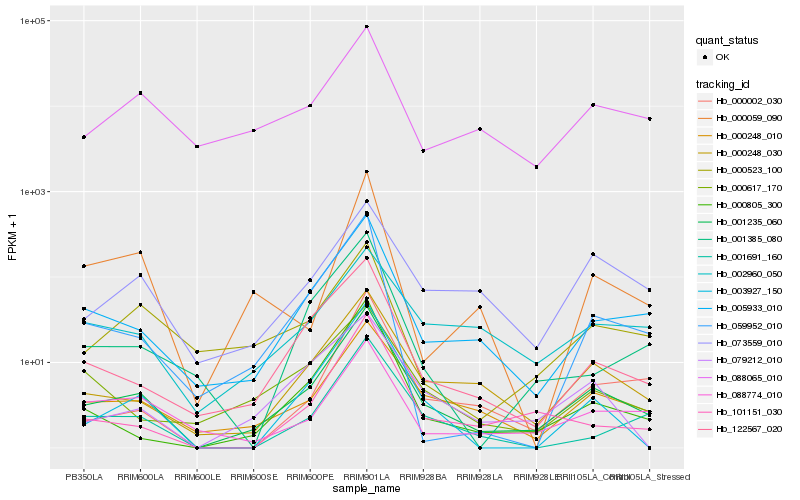
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001235_060 |
0.0 |
- |
- |
- |
| 2 |
Hb_005933_010 |
0.1350763753 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Populus euphratica] |
| 3 |
Hb_000248_030 |
0.1377244435 |
- |
- |
hypothetical protein Pmar_PMAR005331 [Perkinsus marinus ATCC 50983] |
| 4 |
Hb_000248_010 |
0.1428585129 |
- |
- |
hypothetical protein PRUPE_ppa008056mg [Prunus persica] |
| 5 |
Hb_122567_020 |
0.1509662296 |
- |
- |
hypothetical protein CICLE_v10009900mg [Citrus clementina] |
| 6 |
Hb_000805_300 |
0.1622764565 |
- |
- |
- |
| 7 |
Hb_000002_030 |
0.1745952877 |
- |
- |
- |
| 8 |
Hb_003927_150 |
0.1780520276 |
- |
- |
- |
| 9 |
Hb_101151_030 |
0.1816048336 |
- |
- |
- |
| 10 |
Hb_001691_160 |
0.1822513777 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_059952_010 |
0.1822640288 |
- |
- |
PREDICTED: probable protein S-acyltransferase 7 [Eucalyptus grandis] |
| 12 |
Hb_073559_010 |
0.1896735607 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 12 [Jatropha curcas] |
| 13 |
Hb_079212_010 |
0.1897751355 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_088065_010 |
0.1935791409 |
- |
- |
hypothetical protein glysoja_007934 [Glycine soja] |
| 15 |
Hb_088774_010 |
0.195435751 |
- |
- |
- |
| 16 |
Hb_000617_170 |
0.1955732602 |
- |
- |
- |
| 17 |
Hb_001385_080 |
0.1963463873 |
- |
- |
- |
| 18 |
Hb_000523_100 |
0.1974317458 |
- |
- |
PREDICTED: protein DJ-1 homolog D-like [Populus euphratica] |
| 19 |
Hb_002960_050 |
0.2043221899 |
- |
- |
PREDICTED: uncharacterized protein LOC105635325 [Jatropha curcas] |
| 20 |
Hb_000059_090 |
0.2060795453 |
- |
- |
- |