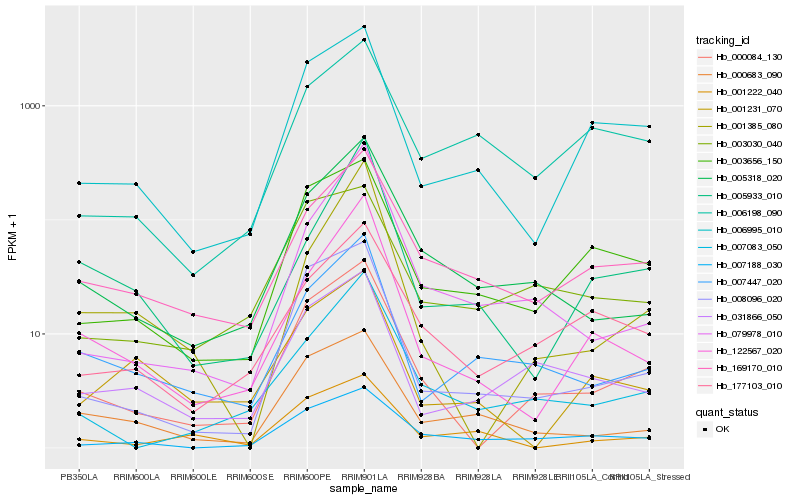
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007447_020 |
0.0 |
- |
- |
hypothetical protein CISIN_1g038065mg [Citrus sinensis] |
| 2 |
Hb_031866_050 |
0.1236648413 |
- |
- |
PREDICTED: uncharacterized protein LOC101756357 [Setaria italica] |
| 3 |
Hb_000683_090 |
0.1310333951 |
- |
- |
stress enhanced protein [Medicago truncatula] |
| 4 |
Hb_005318_020 |
0.1393805022 |
- |
- |
PREDICTED: lecithin-cholesterol acyltransferase-like 4 isoform X3 [Jatropha curcas] |
| 5 |
Hb_169170_010 |
0.1473978227 |
- |
- |
PREDICTED: protein disulfide-isomerase 5-1 isoform X2 [Nelumbo nucifera] |
| 6 |
Hb_122567_020 |
0.1752838217 |
- |
- |
hypothetical protein CICLE_v10009900mg [Citrus clementina] |
| 7 |
Hb_079978_010 |
0.1833666544 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP71 isoform X1 [Nelumbo nucifera] |
| 8 |
Hb_007083_050 |
0.1854451567 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 12-like isoform X2 [Nelumbo nucifera] |
| 9 |
Hb_005933_010 |
0.1857851171 |
- |
- |
PREDICTED: putative disease resistance RPP13-like protein 1 [Populus euphratica] |
| 10 |
Hb_008096_020 |
0.1876245484 |
- |
- |
- |
| 11 |
Hb_000084_130 |
0.1910945798 |
- |
- |
- |
| 12 |
Hb_006995_010 |
0.1934060953 |
- |
- |
PREDICTED: 14 kDa zinc-binding protein [Jatropha curcas] |
| 13 |
Hb_003656_150 |
0.2001467127 |
- |
- |
- |
| 14 |
Hb_001222_040 |
0.2005473776 |
- |
- |
- |
| 15 |
Hb_003030_040 |
0.2007602888 |
- |
- |
hypothetical protein CISIN_1g0466212mg, partial [Citrus sinensis] |
| 16 |
Hb_001231_070 |
0.2043609126 |
- |
- |
- |
| 17 |
Hb_177103_010 |
0.2056155547 |
- |
- |
PREDICTED: histone H2AX [Elaeis guineensis] |
| 18 |
Hb_006198_090 |
0.2075272758 |
- |
- |
PREDICTED: vesicle-associated membrane protein 721-like isoform X2 [Tarenaya hassleriana] |
| 19 |
Hb_007188_030 |
0.2089587534 |
- |
- |
PREDICTED: DNA topoisomerase 3-alpha-like [Malus domestica] |
| 20 |
Hb_001385_080 |
0.2112025139 |
- |
- |
- |