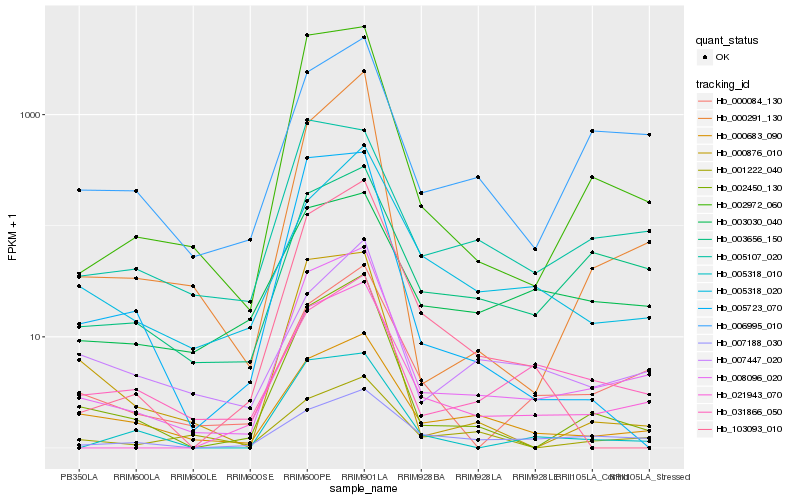
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008096_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_021943_070 |
0.134448206 |
- |
- |
- |
| 3 |
Hb_002450_130 |
0.1365917048 |
- |
- |
hypothetical protein JCGZ_06565 [Jatropha curcas] |
| 4 |
Hb_002972_060 |
0.1403231895 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 5 |
Hb_000084_130 |
0.1487396279 |
- |
- |
- |
| 6 |
Hb_003656_150 |
0.1546278501 |
- |
- |
- |
| 7 |
Hb_006995_010 |
0.1591373524 |
- |
- |
PREDICTED: 14 kDa zinc-binding protein [Jatropha curcas] |
| 8 |
Hb_000683_090 |
0.1599920224 |
- |
- |
stress enhanced protein [Medicago truncatula] |
| 9 |
Hb_005107_020 |
0.1777993367 |
- |
- |
hypothetical protein JCGZ_25266 [Jatropha curcas] |
| 10 |
Hb_005318_020 |
0.1797191472 |
- |
- |
PREDICTED: lecithin-cholesterol acyltransferase-like 4 isoform X3 [Jatropha curcas] |
| 11 |
Hb_000291_130 |
0.1808971421 |
- |
- |
- |
| 12 |
Hb_031866_050 |
0.181311461 |
- |
- |
PREDICTED: uncharacterized protein LOC101756357 [Setaria italica] |
| 13 |
Hb_003030_040 |
0.1844201703 |
- |
- |
hypothetical protein CISIN_1g0466212mg, partial [Citrus sinensis] |
| 14 |
Hb_005318_010 |
0.1850313523 |
- |
- |
hypothetical protein POPTR_0012s15190g, partial [Populus trichocarpa] |
| 15 |
Hb_007188_030 |
0.1864401369 |
- |
- |
PREDICTED: DNA topoisomerase 3-alpha-like [Malus domestica] |
| 16 |
Hb_007447_020 |
0.1876245484 |
- |
- |
hypothetical protein CISIN_1g038065mg [Citrus sinensis] |
| 17 |
Hb_103093_010 |
0.1906616716 |
- |
- |
PREDICTED: wall-associated receptor kinase 4-like [Elaeis guineensis] |
| 18 |
Hb_000876_010 |
0.19479366 |
- |
- |
Uncharacterized protein TCM_022272 [Theobroma cacao] |
| 19 |
Hb_001222_040 |
0.1989211696 |
- |
- |
- |
| 20 |
Hb_005723_070 |
0.199102461 |
- |
- |
hypothetical protein CISIN_1g011514mg [Citrus sinensis] |