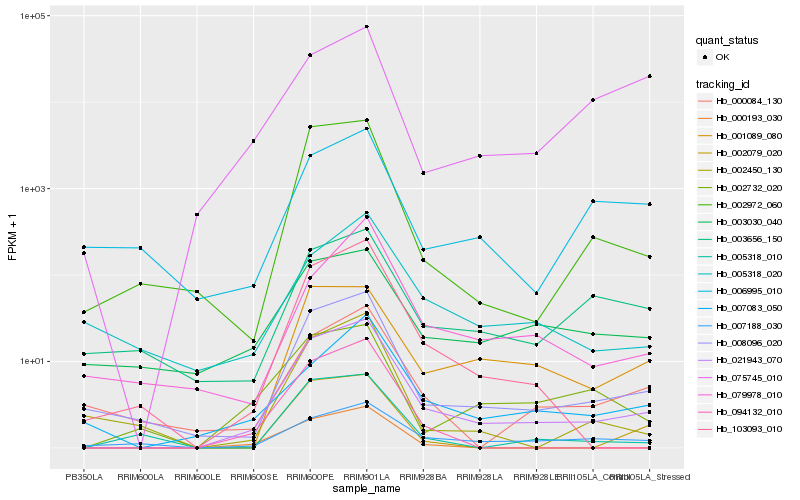
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_021943_070 |
0.0 |
- |
- |
- |
| 2 |
Hb_008096_020 |
0.134448206 |
- |
- |
- |
| 3 |
Hb_002972_060 |
0.1764981558 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 4 |
Hb_103093_010 |
0.1766951469 |
- |
- |
PREDICTED: wall-associated receptor kinase 4-like [Elaeis guineensis] |
| 5 |
Hb_001089_080 |
0.1792096011 |
- |
- |
hypothetical protein glysoja_044047 [Glycine soja] |
| 6 |
Hb_000084_130 |
0.2009262017 |
- |
- |
- |
| 7 |
Hb_002732_020 |
0.2013493093 |
- |
- |
- |
| 8 |
Hb_002450_130 |
0.2015524833 |
- |
- |
hypothetical protein JCGZ_06565 [Jatropha curcas] |
| 9 |
Hb_005318_010 |
0.2016725785 |
- |
- |
hypothetical protein POPTR_0012s15190g, partial [Populus trichocarpa] |
| 10 |
Hb_007188_030 |
0.2027675008 |
- |
- |
PREDICTED: DNA topoisomerase 3-alpha-like [Malus domestica] |
| 11 |
Hb_007083_050 |
0.2037698315 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 12-like isoform X2 [Nelumbo nucifera] |
| 12 |
Hb_003656_150 |
0.2064938158 |
- |
- |
- |
| 13 |
Hb_094132_010 |
0.2078110365 |
- |
- |
hypothetical protein POPTR_0018s03360g [Populus trichocarpa] |
| 14 |
Hb_000193_030 |
0.2128526154 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Jatropha curcas] |
| 15 |
Hb_002079_020 |
0.2136826394 |
- |
- |
- |
| 16 |
Hb_003030_040 |
0.2140420216 |
- |
- |
hypothetical protein CISIN_1g0466212mg, partial [Citrus sinensis] |
| 17 |
Hb_005318_020 |
0.2142059116 |
- |
- |
PREDICTED: lecithin-cholesterol acyltransferase-like 4 isoform X3 [Jatropha curcas] |
| 18 |
Hb_079978_010 |
0.2215151682 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP71 isoform X1 [Nelumbo nucifera] |
| 19 |
Hb_006995_010 |
0.2252129026 |
- |
- |
PREDICTED: 14 kDa zinc-binding protein [Jatropha curcas] |
| 20 |
Hb_075745_010 |
0.229093243 |
- |
- |
hypothetical protein EUGRSUZ_D00579 [Eucalyptus grandis] |