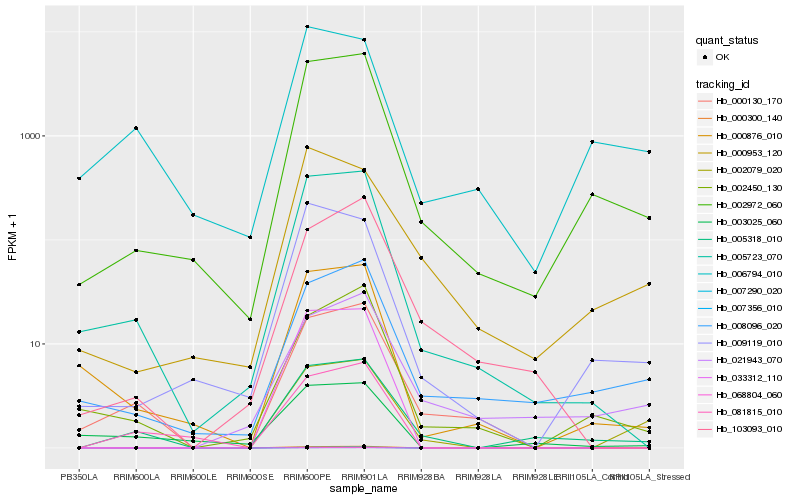
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005318_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0012s15190g, partial [Populus trichocarpa] |
| 2 |
Hb_002972_060 |
0.1512322878 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 3 |
Hb_005723_070 |
0.1820697031 |
- |
- |
hypothetical protein CISIN_1g011514mg [Citrus sinensis] |
| 4 |
Hb_008096_020 |
0.1850313523 |
- |
- |
- |
| 5 |
Hb_000130_170 |
0.2006825556 |
- |
- |
- |
| 6 |
Hb_021943_070 |
0.2016725785 |
- |
- |
- |
| 7 |
Hb_002450_130 |
0.2133806646 |
- |
- |
hypothetical protein JCGZ_06565 [Jatropha curcas] |
| 8 |
Hb_009119_010 |
0.2142837906 |
- |
- |
- |
| 9 |
Hb_103093_010 |
0.2148380629 |
- |
- |
PREDICTED: wall-associated receptor kinase 4-like [Elaeis guineensis] |
| 10 |
Hb_006794_010 |
0.2149802226 |
- |
- |
elicitor-responsive protein [Hevea brasiliensis] |
| 11 |
Hb_000953_120 |
0.222130234 |
- |
- |
- |
| 12 |
Hb_081815_010 |
0.2252107635 |
- |
- |
PREDICTED: uncharacterized protein LOC100266863 [Vitis vinifera] |
| 13 |
Hb_003025_060 |
0.2276585066 |
- |
- |
PREDICTED: protein TOO MANY MOUTHS [Jatropha curcas] |
| 14 |
Hb_002079_020 |
0.2279218203 |
- |
- |
- |
| 15 |
Hb_000876_010 |
0.2307277864 |
- |
- |
Uncharacterized protein TCM_022272 [Theobroma cacao] |
| 16 |
Hb_000300_140 |
0.2355132697 |
transcription factor |
TF Family: HB |
Protein WUSCHEL, putative [Ricinus communis] |
| 17 |
Hb_068804_060 |
0.2360349421 |
- |
- |
PREDICTED: ABC transporter G family member 17-like [Jatropha curcas] |
| 18 |
Hb_007290_020 |
0.2360499963 |
- |
- |
PREDICTED: subtilisin-like protease SBT4.14 [Jatropha curcas] |
| 19 |
Hb_033312_110 |
0.236373226 |
- |
- |
- |
| 20 |
Hb_007356_010 |
0.2366488159 |
- |
- |
CTV.19 [Citrus trifoliata] |