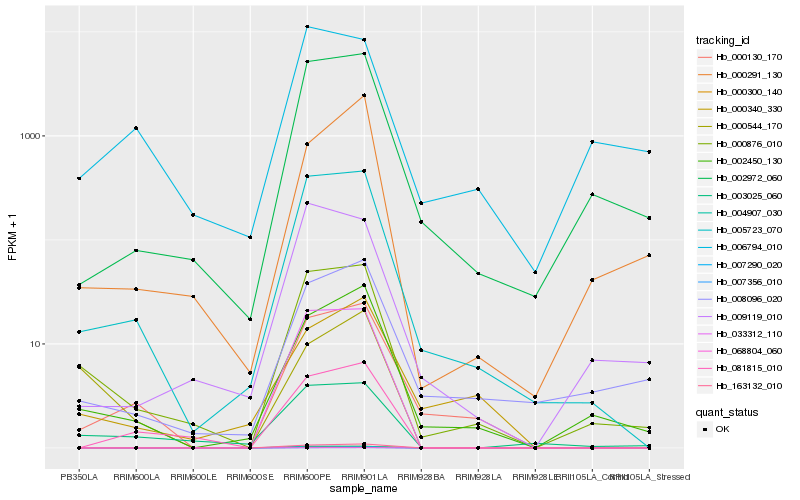
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000876_010 |
0.0 |
- |
- |
Uncharacterized protein TCM_022272 [Theobroma cacao] |
| 2 |
Hb_005723_070 |
0.128804714 |
- |
- |
hypothetical protein CISIN_1g011514mg [Citrus sinensis] |
| 3 |
Hb_002450_130 |
0.1546765159 |
- |
- |
hypothetical protein JCGZ_06565 [Jatropha curcas] |
| 4 |
Hb_002972_060 |
0.1617319272 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 5 |
Hb_003025_060 |
0.1635100536 |
- |
- |
PREDICTED: protein TOO MANY MOUTHS [Jatropha curcas] |
| 6 |
Hb_000130_170 |
0.185328578 |
- |
- |
- |
| 7 |
Hb_009119_010 |
0.1899899726 |
- |
- |
- |
| 8 |
Hb_008096_020 |
0.19479366 |
- |
- |
- |
| 9 |
Hb_000300_140 |
0.2073472951 |
transcription factor |
TF Family: HB |
Protein WUSCHEL, putative [Ricinus communis] |
| 10 |
Hb_033312_110 |
0.2074992398 |
- |
- |
- |
| 11 |
Hb_068804_060 |
0.2080481451 |
- |
- |
PREDICTED: ABC transporter G family member 17-like [Jatropha curcas] |
| 12 |
Hb_007290_020 |
0.2080680203 |
- |
- |
PREDICTED: subtilisin-like protease SBT4.14 [Jatropha curcas] |
| 13 |
Hb_007356_010 |
0.2088482457 |
- |
- |
CTV.19 [Citrus trifoliata] |
| 14 |
Hb_163132_010 |
0.2095561019 |
- |
- |
- |
| 15 |
Hb_004907_030 |
0.2116094429 |
- |
- |
hypothetical protein POPTR_0012s032802g, partial [Populus trichocarpa] |
| 16 |
Hb_006794_010 |
0.2131222881 |
- |
- |
elicitor-responsive protein [Hevea brasiliensis] |
| 17 |
Hb_081815_010 |
0.2137661456 |
- |
- |
PREDICTED: uncharacterized protein LOC100266863 [Vitis vinifera] |
| 18 |
Hb_000291_130 |
0.2166561114 |
- |
- |
- |
| 19 |
Hb_000544_170 |
0.224947314 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000340_330 |
0.2283679667 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.6 [Jatropha curcas] |