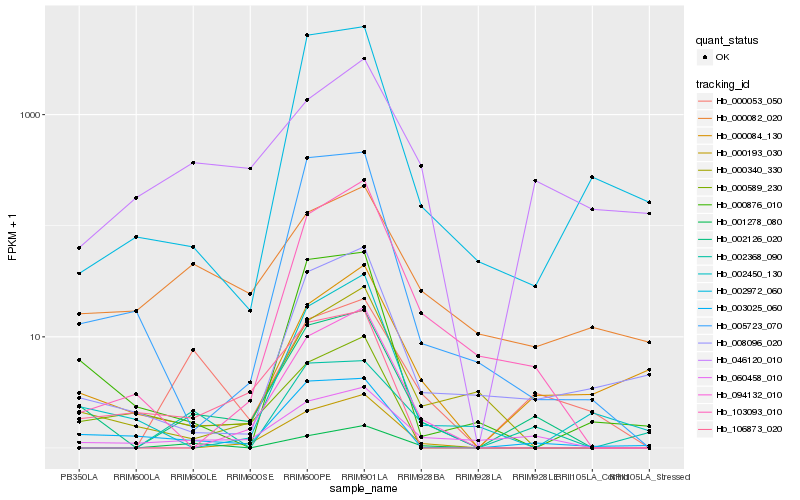
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002126_020 |
0.0 |
- |
- |
ferrochelatase [Medicago truncatula] |
| 2 |
Hb_060458_010 |
0.1890963735 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003025_060 |
0.195433408 |
- |
- |
PREDICTED: protein TOO MANY MOUTHS [Jatropha curcas] |
| 4 |
Hb_106873_020 |
0.2257892474 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 5 |
Hb_005723_070 |
0.231761113 |
- |
- |
hypothetical protein CISIN_1g011514mg [Citrus sinensis] |
| 6 |
Hb_000876_010 |
0.2340349881 |
- |
- |
Uncharacterized protein TCM_022272 [Theobroma cacao] |
| 7 |
Hb_000193_030 |
0.2347854975 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Jatropha curcas] |
| 8 |
Hb_103093_010 |
0.23665313 |
- |
- |
PREDICTED: wall-associated receptor kinase 4-like [Elaeis guineensis] |
| 9 |
Hb_094132_010 |
0.2387790685 |
- |
- |
hypothetical protein POPTR_0018s03360g [Populus trichocarpa] |
| 10 |
Hb_000340_330 |
0.2464522972 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.6 [Jatropha curcas] |
| 11 |
Hb_000589_230 |
0.2476731256 |
- |
- |
unknown [Picea sitchensis] |
| 12 |
Hb_008096_020 |
0.248623284 |
- |
- |
- |
| 13 |
Hb_002972_060 |
0.256106129 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 14 |
Hb_000084_130 |
0.2575775322 |
- |
- |
- |
| 15 |
Hb_001278_080 |
0.2606529244 |
transcription factor |
TF Family: AUX/IAA |
hypothetical protein JCGZ_09311 [Jatropha curcas] |
| 16 |
Hb_002450_130 |
0.2608057948 |
- |
- |
hypothetical protein JCGZ_06565 [Jatropha curcas] |
| 17 |
Hb_046120_010 |
0.2630550597 |
- |
- |
- |
| 18 |
Hb_000053_050 |
0.2630562162 |
- |
- |
hypothetical protein F775_01878 [Aegilops tauschii] |
| 19 |
Hb_002368_090 |
0.2637915264 |
- |
- |
PREDICTED: cationic peroxidase 1-like [Populus euphratica] |
| 20 |
Hb_000082_020 |
0.2647467154 |
- |
- |
- |