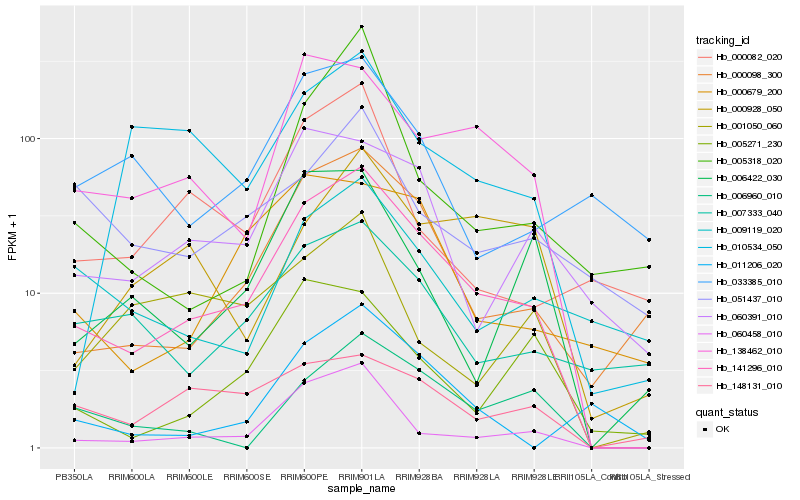
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_141296_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_060458_010 |
0.1702009413 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000098_300 |
0.1717974428 |
- |
- |
- |
| 4 |
Hb_138462_010 |
0.1868421197 |
- |
- |
PREDICTED: SUMO-conjugating enzyme UBC9-A-like isoform X1 [Elaeis guineensis] |
| 5 |
Hb_060391_010 |
0.2145522926 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 6 |
Hb_007333_040 |
0.2246161918 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK1-like [Glycine max] |
| 7 |
Hb_006960_010 |
0.2257007446 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000679_200 |
0.2296525657 |
- |
- |
EIN3-binding F box protein 1 [Theobroma cacao] |
| 9 |
Hb_148131_010 |
0.2314879298 |
- |
- |
PREDICTED: ribosome biogenesis protein WDR12-like, partial [Jatropha curcas] |
| 10 |
Hb_009119_020 |
0.2355899045 |
- |
- |
hypothetical protein CISIN_1g033708mg [Citrus sinensis] |
| 11 |
Hb_006422_030 |
0.2361228805 |
- |
- |
- |
| 12 |
Hb_005271_230 |
0.2421099119 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X4 [Jatropha curcas] |
| 13 |
Hb_000082_020 |
0.2424934094 |
- |
- |
- |
| 14 |
Hb_011206_020 |
0.2457323362 |
- |
- |
- |
| 15 |
Hb_000928_050 |
0.2473857515 |
- |
- |
PREDICTED: DNA polymerase delta subunit 4 [Jatropha curcas] |
| 16 |
Hb_033385_010 |
0.2479913011 |
- |
- |
hypothetical protein JCGZ_06094 [Jatropha curcas] |
| 17 |
Hb_005318_020 |
0.2487100368 |
- |
- |
PREDICTED: lecithin-cholesterol acyltransferase-like 4 isoform X3 [Jatropha curcas] |
| 18 |
Hb_010534_050 |
0.2498305372 |
- |
- |
PREDICTED: 40S ribosomal protein S7 [Jatropha curcas] |
| 19 |
Hb_051437_010 |
0.2532842566 |
- |
- |
PREDICTED: calmodulin-interacting protein 111 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001050_060 |
0.2604093957 |
- |
- |
hypothetical protein POPTR_0011s036602g, partial [Populus trichocarpa] |