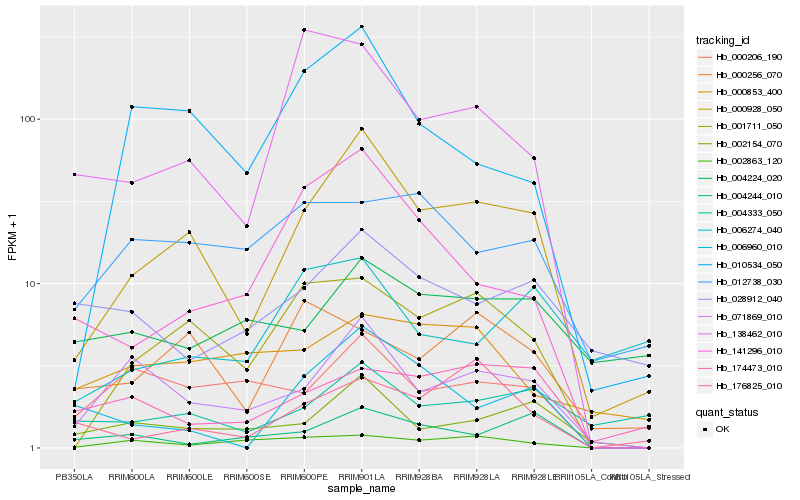
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000928_050 |
0.0 |
- |
- |
PREDICTED: DNA polymerase delta subunit 4 [Jatropha curcas] |
| 2 |
Hb_001711_050 |
0.2049843251 |
- |
- |
- |
| 3 |
Hb_071869_010 |
0.2084050984 |
- |
- |
PREDICTED: uncharacterized protein LOC105628378 isoform X2 [Jatropha curcas] |
| 4 |
Hb_002154_070 |
0.2139255647 |
desease resistance |
Gene Name: AAA_25 |
hypothetical protein Csa_6G014750 [Cucumis sativus] |
| 5 |
Hb_010534_050 |
0.2349415873 |
- |
- |
PREDICTED: 40S ribosomal protein S7 [Jatropha curcas] |
| 6 |
Hb_006960_010 |
0.2421846601 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_141296_010 |
0.2473857515 |
- |
- |
- |
| 8 |
Hb_004333_050 |
0.2481910236 |
- |
- |
- |
| 9 |
Hb_138462_010 |
0.2591843117 |
- |
- |
PREDICTED: SUMO-conjugating enzyme UBC9-A-like isoform X1 [Elaeis guineensis] |
| 10 |
Hb_000206_190 |
0.2593868269 |
- |
- |
PREDICTED: F-box/LRR-repeat protein At4g14103-like [Pyrus x bretschneideri] |
| 11 |
Hb_004244_010 |
0.2625640331 |
- |
- |
hypothetical protein JCGZ_06191 [Jatropha curcas] |
| 12 |
Hb_176825_010 |
0.268885883 |
- |
- |
PREDICTED: uncharacterized protein LOC105630857 isoform X2 [Jatropha curcas] |
| 13 |
Hb_174473_010 |
0.2701877047 |
- |
- |
PREDICTED: uncharacterized protein LOC105775505 [Gossypium raimondii] |
| 14 |
Hb_028912_040 |
0.271173219 |
- |
- |
hypothetical protein POPTR_0016s03250g [Populus trichocarpa] |
| 15 |
Hb_000256_070 |
0.2716274549 |
- |
- |
PREDICTED: uncharacterized protein At2g39795, mitochondrial [Jatropha curcas] |
| 16 |
Hb_012738_030 |
0.2775257819 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002863_120 |
0.2827441226 |
desease resistance |
Gene Name: AAA_17 |
JHL25H03.3 [Jatropha curcas] |
| 18 |
Hb_004224_020 |
0.2829247153 |
- |
- |
PREDICTED: uncharacterized protein LOC105643786 [Jatropha curcas] |
| 19 |
Hb_006274_040 |
0.2848068783 |
- |
- |
PREDICTED: DENN domain and WD repeat-containing protein SCD1-like [Fragaria vesca subsp. vesca] |
| 20 |
Hb_000853_400 |
0.2849926073 |
- |
- |
PREDICTED: CTP synthase [Jatropha curcas] |