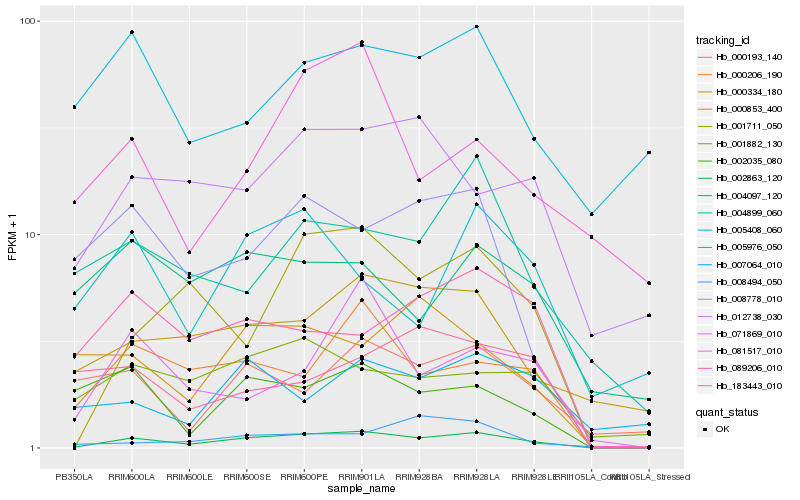
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002863_120 |
0.0 |
desease resistance |
Gene Name: AAA_17 |
JHL25H03.3 [Jatropha curcas] |
| 2 |
Hb_000206_190 |
0.1997114689 |
- |
- |
PREDICTED: F-box/LRR-repeat protein At4g14103-like [Pyrus x bretschneideri] |
| 3 |
Hb_001711_050 |
0.2000464036 |
- |
- |
- |
| 4 |
Hb_000853_400 |
0.2220029479 |
- |
- |
PREDICTED: CTP synthase [Jatropha curcas] |
| 5 |
Hb_008778_010 |
0.2242428545 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_002035_080 |
0.2278382362 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_001882_130 |
0.2296576849 |
transcription factor |
TF Family: mTERF |
hypothetical protein VITISV_028996 [Vitis vinifera] |
| 8 |
Hb_071869_010 |
0.229979877 |
- |
- |
PREDICTED: uncharacterized protein LOC105628378 isoform X2 [Jatropha curcas] |
| 9 |
Hb_012738_030 |
0.2320759034 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_089206_010 |
0.2323516624 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 11 |
Hb_004899_060 |
0.2405576769 |
- |
- |
46 kDa ketoavyl-ACP synthase [Ricinus communis] |
| 12 |
Hb_000334_180 |
0.2457950759 |
- |
- |
hypothetical protein JCGZ_16779 [Jatropha curcas] |
| 13 |
Hb_000193_140 |
0.2461313088 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_005408_060 |
0.2527181723 |
- |
- |
PREDICTED: uncharacterized protein LOC105648045 isoform X1 [Jatropha curcas] |
| 15 |
Hb_081517_010 |
0.2561634413 |
- |
- |
PREDICTED: putative potassium transporter 12 isoform X2 [Jatropha curcas] |
| 16 |
Hb_008494_050 |
0.2578770159 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 17 |
Hb_004097_120 |
0.2580011131 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_007064_010 |
0.2610819034 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39680 [Jatropha curcas] |
| 19 |
Hb_183443_010 |
0.2615238119 |
- |
- |
PREDICTED: uncharacterized protein LOC105786773 [Gossypium raimondii] |
| 20 |
Hb_005976_050 |
0.2620180408 |
- |
- |
PREDICTED: protein RALF-like 34 [Jatropha curcas] |