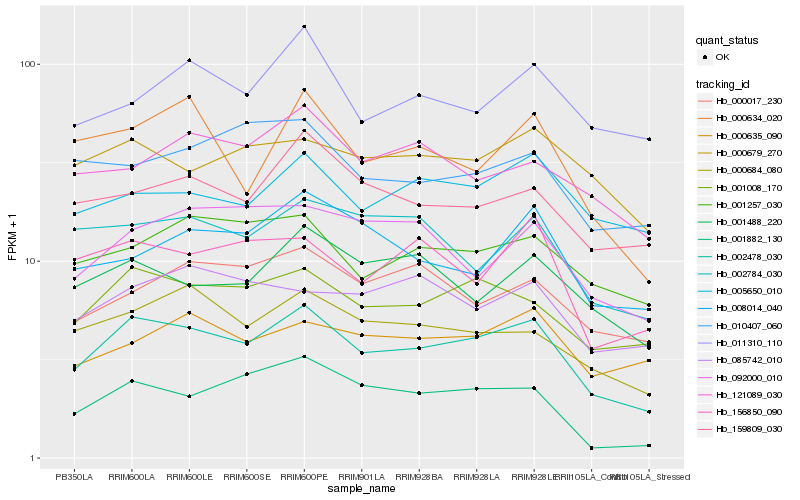
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002478_030 |
0.0 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 1-like [Jatropha curcas] |
| 2 |
Hb_001008_170 |
0.0917248485 |
transcription factor |
TF Family: mTERF |
RNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_000684_080 |
0.1059024733 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.1 [Jatropha curcas] |
| 4 |
Hb_001257_030 |
0.1138980431 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek5 [Jatropha curcas] |
| 5 |
Hb_159809_030 |
0.1161899156 |
- |
- |
PREDICTED: UDP-D-xylose:L-fucose alpha-1,3-D-xylosyltransferase 2-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_001488_220 |
0.1180118087 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 7 |
Hb_000634_020 |
0.1192330439 |
- |
- |
hypothetical protein JCGZ_11888 [Jatropha curcas] |
| 8 |
Hb_156850_090 |
0.1200822154 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 9 |
Hb_002784_030 |
0.1211100692 |
- |
- |
hypothetical protein POPTR_0001s06740g [Populus trichocarpa] |
| 10 |
Hb_000017_230 |
0.1215955098 |
- |
- |
hypothetical protein JCGZ_20558 [Jatropha curcas] |
| 11 |
Hb_005650_010 |
0.1219044713 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 12 |
Hb_000679_270 |
0.1226840337 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |
| 13 |
Hb_001882_130 |
0.123929842 |
transcription factor |
TF Family: mTERF |
hypothetical protein VITISV_028996 [Vitis vinifera] |
| 14 |
Hb_008014_040 |
0.1239814168 |
- |
- |
PREDICTED: uncharacterized protein LOC105649721 [Jatropha curcas] |
| 15 |
Hb_011310_110 |
0.1240858133 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At5g25400 isoform X1 [Jatropha curcas] |
| 16 |
Hb_121089_030 |
0.1246716971 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 17 |
Hb_092000_010 |
0.1249524606 |
- |
- |
kelch repeat-containing F-box family protein [Populus trichocarpa] |
| 18 |
Hb_000635_090 |
0.1252965363 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 19 |
Hb_085742_010 |
0.1266165456 |
- |
- |
hypothetical protein POPTR_0005s05170g [Populus trichocarpa] |
| 20 |
Hb_010407_060 |
0.1270867407 |
- |
- |
PREDICTED: probable methyltransferase PMT3 [Jatropha curcas] |