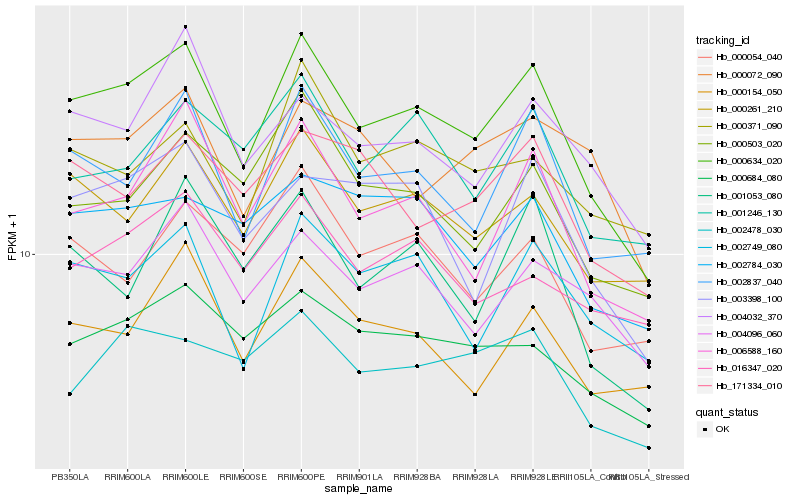
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000634_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_11888 [Jatropha curcas] |
| 2 |
Hb_002837_040 |
0.0930422834 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 3 |
Hb_006588_160 |
0.096351308 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic isoform X2 [Jatropha curcas] |
| 4 |
Hb_000261_210 |
0.1011322266 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 5 |
Hb_002749_080 |
0.1018401387 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000684_080 |
0.108559344 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.1 [Jatropha curcas] |
| 7 |
Hb_004096_060 |
0.1086250317 |
- |
- |
PREDICTED: probable dolichyl pyrophosphate Man9GlcNAc2 alpha-1,3-glucosyltransferase isoform X2 [Jatropha curcas] |
| 8 |
Hb_003398_100 |
0.1113398699 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000154_050 |
0.1146923122 |
- |
- |
CMP-sialic acid transporter, putative [Ricinus communis] |
| 10 |
Hb_000503_020 |
0.1148204458 |
- |
- |
PREDICTED: calcium-dependent protein kinase 13 [Jatropha curcas] |
| 11 |
Hb_001053_080 |
0.1155223823 |
- |
- |
OsCesA3 protein [Morus notabilis] |
| 12 |
Hb_001246_130 |
0.1176389504 |
- |
- |
PREDICTED: NAD-dependent malic enzyme 62 kDa isoform, mitochondrial [Jatropha curcas] |
| 13 |
Hb_004032_370 |
0.117920614 |
- |
- |
pyrophosphate-dependent phosphofructokinase [Hevea brasiliensis] |
| 14 |
Hb_002478_030 |
0.1192330439 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 1-like [Jatropha curcas] |
| 15 |
Hb_000054_040 |
0.1197833223 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1 [Jatropha curcas] |
| 16 |
Hb_002784_030 |
0.1201149832 |
- |
- |
hypothetical protein POPTR_0001s06740g [Populus trichocarpa] |
| 17 |
Hb_171334_010 |
0.1203704084 |
- |
- |
PREDICTED: CDPK-related kinase 5 [Jatropha curcas] |
| 18 |
Hb_016347_020 |
0.1227025217 |
- |
- |
PREDICTED: D-amino-acid transaminase, chloroplastic-like isoform X1 [Populus euphratica] |
| 19 |
Hb_000371_090 |
0.1234959915 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit-like [Jatropha curcas] |
| 20 |
Hb_000072_090 |
0.124834794 |
- |
- |
PREDICTED: SEC14 cytosolic factor [Jatropha curcas] |